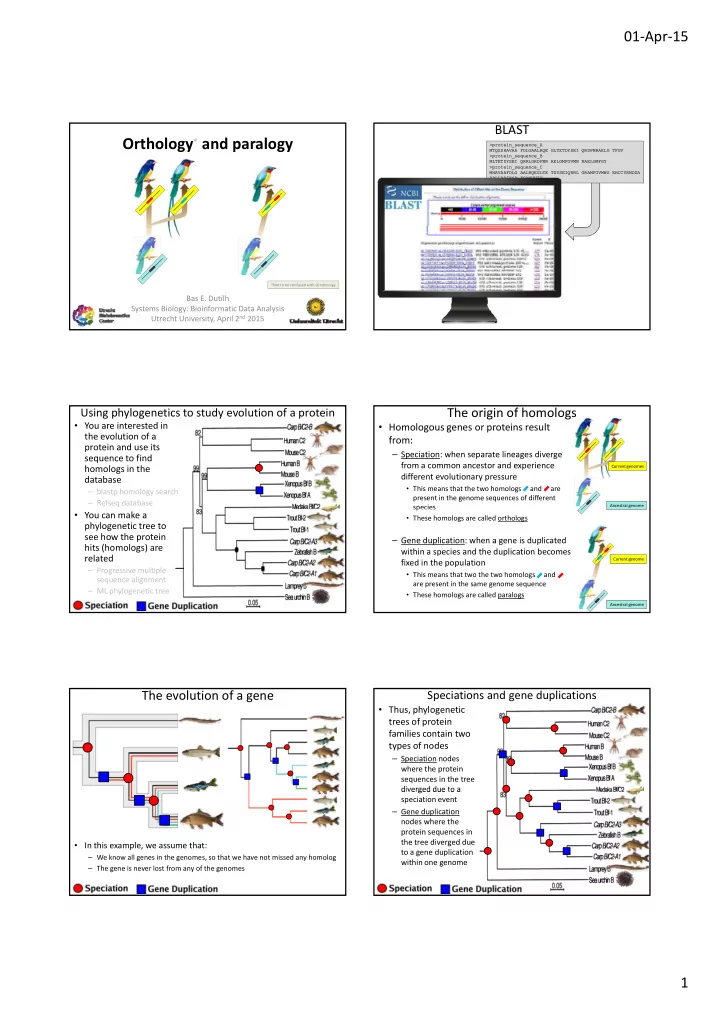

01 ‐ Apr ‐ 15 BLAST Orthology * and paralogy >pro >p rote tein in_s _seq eque uence_A MTQSSHAVAA FD FDLGA LGAAL ALRQ RQE GL E GLTET TETDY DYSE SEI QR I QRDP DPNR NRAEL AELG TF G TFGV GV >pro >p rote tein in_s _seq eque uence_B MLTETDYSEI QR QRRLG RLGRD RDPN PNR AE R AELGM LGMFG FGVM VMN RA N RAEL ELGM GMFGY FGY >p >pro rote tein in_s _seq eque uence_C MHAVAAFDLG AA AALRQ LRQEG EGLT LTE TD E TDYSE YSEIQ IQRR RRL GR L GRAM AMFG FGVMW VMWS EH S EHCC CCYR YRNDD NDDA RP RPLL LLRP RPIK IKSP SP F FGAWVVIV *Not to be confused with Ornithology Bas E. Dutilh Systems Biology: Bioinformatic Data Analysis Utrecht University, April 2 nd 2015 Using phylogenetics to study evolution of a protein The origin of homologs • You are interested in • Homologous genes or proteins result the evolution of a from: protein and use its – Speciation: when separate lineages diverge sequence to find from a common ancestor and experience homologs in the Current genomes different evolutionary pressure database • This means that the two homologs and are – blastp homology search present in the genome sequences of different – Refseq database species species Ancestral genome • You can make a • These homologs are called orthologs phylogenetic tree to see how the protein – Gene duplication: when a gene is duplicated hits (homologs) are within a species and the duplication becomes related Current genome fixed in the population – Progressive multiple • This means that two the two homologs and sequence alignment are present in the same genome sequence – ML phylogenetic tree • These homologs are called paralogs Ancestral genome The evolution of a gene Speciations and gene duplications • Thus, phylogenetic trees of protein families contain two types of nodes – Speciation nodes where the protein sequences in the tree diverged due to a d e ged due o a speciation event – Gene duplication nodes where the protein sequences in the tree diverged due • In this example, we assume that: to a gene duplication – We know all genes in the genomes, so that we have not missed any homolog within one genome – The gene is never lost from any of the genomes 1
01 ‐ Apr ‐ 15 How to tell the difference? The evolution of gene functions • Orthologs are directly related to the • If the two daughter branches of a node same ancestral gene, but have been contain the same retained in different species species, it is a gene – Since there is only one gene to perform the Current genomes duplication node function, orthologous proteins generally perform the same function in these different – Only if those two genomes proteins are both really present in the same Ancestral genome genome and there has genome and there has • Paralogs have evolved side by side in the been no horizontal gene same genome for a while transfer (HGT) – Since there are two genes with a redundant • Otherwise it can be a function, paralogous proteins might be more speciation node free to change their function in time Current genome – Unless genes were lost – Some can acquire new functions (neo ‐ and or missed in these sub ‐ functionalization) genomes – this is called unrecognized paralogy – Most gene families evolve through duplications Ancestral genome Question Answers Mouse A Mouse C Mouse A Mouse C Human A Mouse D Human A Mouse D Human C Human C Mouse B Mouse B Human D Human D Human B Human B Tree 1 Tree 2 Tree 1 Tree 2 • Observe the two simplified gene trees above of two • Observe the two simplified gene trees above of two homologs from mouse and two homologs from human homologs from mouse and two homologs from human. homologs from mouse and two homologs from human. homologs from mouse and two homologs from human. 1. See above: 1. Which are the speciation nodes and which are the gene duplication nodes? 2. The Mouse A and Human A genes in Tree 1 are orthologs. 3. The Human C and Human D genes in Tree 2 are paralogs. 2. What kind of homologs are Mouse A and Human A in Tree 1? 4. In Tree 1, Mouse A / Human A , and Mouse B / Human B are 3. What kind of homologs are Human C and Human D in Tree 2? more likely to have the same function. 4. Which genes may have the same function in Tree 1? 5. We cannot say which genes may have the same function in Tree 5. Which genes may have the same function in Tree 2? 2. What we can say is that the genes are all homologous, so they will probably have a similar function to some extent. Using orthology for function prediction Identifying orthologs • Orthologs are best identified by studying phylogenetic trees – They are derived from a speciation node – There can be simple 1:1 orthologs, but if one or both of the daughter lineages expanded by gene duplication (possibly resulting in many paralogs), there can also be 1:many or many:many orthology relationships Present species mouse human human mouse Hex2 Hex1 Hex1 Hex1 Hex2 Hex2 – This can make function prediction based on orthology difficult • Operational definition of orthology: bi ‐ directional best hits (BBH) Time mouse _ Hex1 is the best hit – Blast human _ Hex1 against all proteins in mouse Protein_1 Sp_2 Protein_2 Hex1 Hex2 Ancestor – Blast mouse Hex1 against all proteins in human if human Hex1 is the best hit, Blast mouse _ Hex1 against all proteins in human if human _ Hex1 is the best hit, Sp_1 p_ Protein 2 _ Protein 1 _ of the then human _ Hex1 and mouse _ Hex1 are probably orthologs Protein_1 Protein_2 tetrapods – If there is no 1:1 BBH, then it is likely that one or both of the orthologs duplicated Hex since the speciation event • Researchers are often trying to identify orthologs in model frog mouse human human mouse frog organisms, because if the function of the ortholog has been studied Hex2 Hex1 Hex1 Hex1 Hex2 Hex2 in the model organism, it might perform the same function in other organisms as well Hex1 Hex2 • However, note that the definition of orthology says nothing about function Hex 2
01 ‐ Apr ‐ 15 Gene losses Loss in the ancestor of Medaka and Trout • If we assume that this tree contains all the members of this gene family in the species shown, then we can deduce where gene losses occurred: • How many copies of y p this gene did the last common ancestor of all fishes have? • How many copies of this gene did the last common ancestor of all mammals have? 14 Gene loss Gene evolution can quickly get very complex The “Tree” of Life? • Complex evolutionary processes plus several endosymbiosis species A events such as the mitochondrion ancestor and the chloroplast, have led to species B an intricate network of Gene invention evolutionary relationships that Speciation node (orthologs) might not best be represented in Gene duplication node (paralogs) species C the form of a Tree of Life Gene loss • There are many possible causes of conflict between the phylogenetic tree of a gene and the species tree or “Tree of Life”: – Unrecognized paralogy after differential gene losses – Horizontal gene transfer – Mutation saturation, biases, different rates of evolution Detecting HGT in trees Detecting HGT in trees 3
Recommend
More recommend