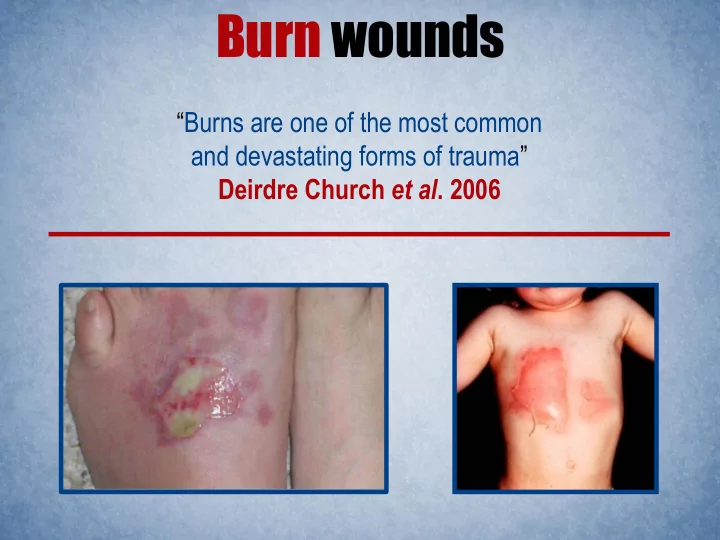

Burn wounds “ Burns are one of the most common and devastating forms of trauma ” Deirdre Church et al . 2006
Burn wounds 1,200,000 burn injuries 100,000 hospitalized 5,000 deaths 75% are infection related National Center for Injury Prevention and Control in the United States
iGEM Groningen 2014 the smart bandage
Conventional treatments Skin transplantation Crèmes and bandages Preventive antibiotics Bathing
Requirements the smart bandage • Must be used for several days • Must detect infections • Must secrete infection preventing molecules (IPMs)
Opportunistic pathogens Staphylococcus aureus Pseudomonas aeruginosa Gram-positive Gram-negative
“A post -antibiotic era - in which common infections and minor injuries can kill - far from being an apocalyptic fantasy, is instead a very real possibility for the 21st Century.” WHO, April 2014 World Health Organization (2014) Antimicrobial resistance: global report on surveillance. ISBN 978 92 4 156474 8
Our goal To detect and prevent further infections caused by S. aureus and P. aeruginosa in burn wounds.
Why Lactococcus lactis ? • Harmless species • Food approved • Produces lactate • No spore formation • Can be temporarily inactivated Image adapted from a SEM scan by Joseph A. Heintz, University of Wisconsin-Madison.
Commission of Genetic Modification Thomas and Lianne at COGEM.
How does it work? DETECTION Quorum sensing molecules AHL ( P. aeruginosa ) AIP-1 ( S. aureus ) Contreras, G. et al. (2013) LaSarre, B. and Michael J.F. (2013)
How does it work? BIOFILM DESTRUCTION DspB degrades the biofilms of both species This way the other infection prevention molecules can reach them Mark, B.L. et al. (2001)
How does it work? QUORUM SENSING DISRUPTION AiiA inhibits AHL Quorum sensing of P. aeruginosa is disrupted P. aeruginosa expresses less virulence genes Body can clear P. aeruginosa without problems Kim, M.H. et al. (2005)
How does it work? NISIN PRODUCTION Nisin kills Gram-positive bacteria Wiedemann, I. et al. (2001)
Biobricks Toolbox Nisin biobricks • sfGFP • LactoAid AiiA • DspB • • Gene constructs
From bacterium to bandage
Bandage materials Top layer - polymethyl pentane • Permeable to gases iGEM 2012, Groningen •
Bandage materials Middle layer – polyacrylamide gel • Pore size is adjustable Nutrients can be added • Cheap • Rehydratable •
Bandage materials Bottom layer – cellulose nitrate • Permeable to small molecules and proteins iGEM 2013, TU Delft •
Description of the model Single unit Total model
Parameters – Rate Equations Nisin production assay Optimal concentration 2% Glucose
Models
Model-based design Least optimal design (0 - 24 h) No IPMs Max IPMs
Model-based design Best design (0 - 24 h) No IPMs Max IPMs
Modeling outcome Time to reach effective concentrations nisin aiiA dspB 12 min 18 min 18 min 24 min 30 min 30 min 114 min 138 min 144 min
Modeling outcome Time to reach effective concentrations nisin aiiA dspB 12 min 18 min 18 min 24 min 30 min 30 min 114 min 138 min 144 min
Modeling outcome Time to reach effective concentrations nisin aiiA dspB 12 min 18 min 18 min 24 min 30 min 30 min 114 min 138 min 144 min
Results: Cell growth in the active layer
Results: Nisin Secretion • Nisin-sensitive strain plated • LactoAid active layer, with nisin-producing strain
In conclusion “… this is an application that appeals, it would be a waste if this idea would remain at -80 ° C…”
We would like to thank:
Thank YOU for your attention!
References Wiedemann , I et al. ����. �“pe�ifi� Bi�di�g of Nisi� to the Peptidogl��a� Pre�ursor Lipid II Co��i�es Pore For�atio� a�d I�hi�itio� of Cell Wall Bios��thesis for Pote�t A�ti�ioti� A�tivit�.� The Journal of biological chemistry 276(3): 1772 – 79. Kim, Myung Hee et al. 2005. �The Mole�ular “tru�ture a�d Catal�ti� Me�ha�is� of a Quoru� - Quenching N-Acyl-L-Homoserine La�to�e H�drolase.� Proceedings of the National Academy of Sciences of the United States of America 102(49): 17606 – 11. Mark, B L et al. 2001. �Cr�stallographi� Evide��e for “u�strate -Assisted Catalysis in a Bacterial Beta- Hexosaminidase .� The Journal of biological chemistry 276(13): 10330 – 37. LaSarre, Breah, and Michael J Federle . ����. �E�ploiti�g Quoru� “e�si�g to Co�fuse Ba�terial Pathoge�s.� Microbiology and molecular biology reviews : MMBR 77(1): 73 – 111. García-Contreras, Rodolfo, Toshinari Maeda, a�d Tho�as K Wood. ����. �Resista��e to Quoru� - Que��hi�g Co�pou�ds.� Applied and environmental microbiology 79(22): 6840 – 46.
Rate equations X 1 = X 2 = X 3 = x 7 = X 8 = X 9 = X 10 = X 11 = IPMs = M. Boonmee et al . Biochemical Engineering Journal 14 (2003) 127 – 135 Shimizu et al . Appl. Environ. Microbiol. 1999, 65(7):3134.
Rate equation Rate = F ( variable,parameters) + Diffusion term
Diffusion equations Volume fraction of the gel: m w - mass of the water m b - mass of the buffer m p - mass of the polymer v p - partial specific volume of gel in water v wb - partial specific volume of gel in buffer Density of IPMs: Tong, Jane, and John L. Anderson. Biophysical journal 70.3 (1996): 1505-1513. Fischer Hannes, et al . Protein Science 13.10 (2004): 2825-2828.
Diffusion equations Stokes-Einstein equation: Diffusion rates of the IPMs were found with: Tong, Jane, and John L. Anderson. Biophysical journal 70.3 (1996): 1505-1513.
Model-based design Expected most optimal design No IPMs Max IPMs
Parameters – Rate Equations Growth rate – L. lactis 10 Log OD 600nm 1 0.1 0.01 0 2 3 4 6 7 8 9 10 11 Time (Hours) Growth rate (0.653 hr -1 ) 39 minutes
Rate equation F ( variable,parameters) Rate Diffusion term + =
Results: Protein production in the gel 30 min 60 min 90 min In the Gel On the Gel
Toolbox Nisin gene cluster
Nisin production
Recommend
More recommend