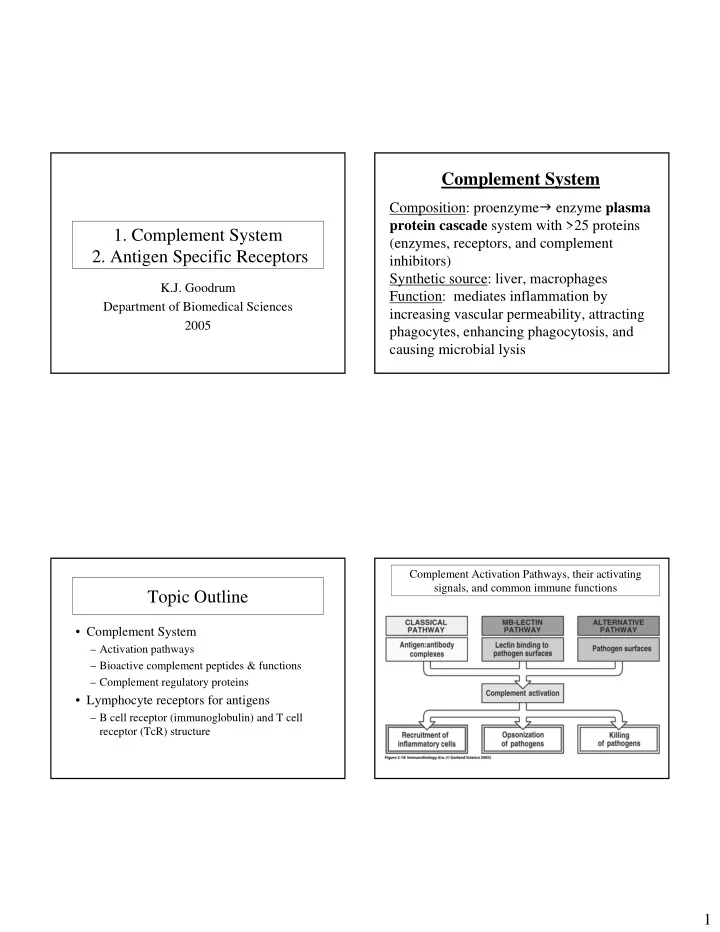

Complement System Composition: proenzyme � enzyme plasma protein cascade system with >25 proteins 1. Complement System (enzymes, receptors, and complement 2. Antigen Specific Receptors inhibitors) Synthetic source: liver, macrophages K.J. Goodrum Function: mediates inflammation by Department of Biomedical Sciences increasing vascular permeability, attracting 2005 phagocytes, enhancing phagocytosis, and causing microbial lysis Complement Activation Pathways, their activating signals, and common immune functions Topic Outline • Complement System – Activation pathways – Bioactive complement peptides & functions – Complement regulatory proteins • Lymphocyte receptors for antigens – B cell receptor (immunoglobulin) and T cell receptor (TcR) structure 1
Classical Pathway Lectin Pathway Alternative Pathway Bioactive Complement Peptides Ag:Ab complex MBL:microorganism C3 H 2 O complex • Plasma protease cascade generates peptides C1q MASP1 C3H 2 O C1r active in inflammation and innate immunity MASP2 B C1s Ba – C3a & C5a anaphylatoxins release histamine from D MBL-MASP1-MASP2 C1 mast cells � � vascular permeability C3(H 2 O)Bb C4 C4a C4 C4a – C5a chemotaxin , recruits phagocytes C3 C3a C14b MC4b Target bound C3b – C3b & C3bi opsonins (coats microbes to enhance C2 C2a C2 C2a B recognition & uptake by phagocytes) C14b2b Ba MC4b2b D – C5b6789n membrane attack complex (lyses C3bBb C3 microbial cell membranes) P C3bBbP C3a + C3b Terminal complement C3 convertases Classical Complement Pathway C3 pathway common to C14b2b all activation MC4b2b C3a pathways C3bBbP C14b2b3b or MC4b2b3b or (C3b)nBbP C5 convertases C5 C5a C5b C6 +C7 C5b67 C8 +C9 C5b678(9)n Membrane attack complex 2
Alternative Complement Pathway -2 Classical Complement Pathway- continued Alternative Complement Pathway -3 Alternative Complement Pathway -1 3
Classical and Alternate Cells detect and Pathway formation of respond to C5 convertase and complement cleavage of C5 peptides via cell-membrane receptors for complement peptides. Terminal Complement Pathway and formation of the Membrane Spontaneous or chronic activation of complement is prevented Attack Complex by inhibitory proteins, both soluble and membrane associated. 4
Antigen Recognition Basic antibody structure = 4 covalently • B cells recognize antigens via membrane attached associated immunoglobulin molecules polypeptide (antibodies) chains with 2 – Antigen-activated B cells synthesize and secrete a soluble form of identical heavy this receptor (plasma antibody) which accumulates in fluids and (H) and 2 specifically binds and eliminates the stimulating antigen. identical light • T cells recognize antigens via a membrane associated chains (L) heterodimeric protein, the T cell receptor or TcR – TcR are not secreted Each H and L chain Antibody Activity have structurally constant (C) and structurally variable (V) • Antibodies specifically recognize and bind domains. to the inducing antigen • Humoral immunity: antibody-mediated immunity can be transferred from an immune to a non-immune person by transfer of serum • The gamma-globulin protein fraction of serum contains the antibody activity 5
Hypervariable amino acid residues in the H The Fab fragment and L chain V regions physically bind the of antibody antigen and determine the specificity of the molecules antigen receptor. contains the antigen binding region. The Fc fragment determines antibody class and effector functions (complement interaction, opsonic activity) The Hypervariable “immunoglobulin amino acid fold” refers to the residues are characteristic 3D located at the structure of V and C exposed ends region domains. of V domain loops. 6
V H and V L domains fold to form antigen binding sites that resemble pockets, grooves, open faces, or extrusions. The structure of the T cell receptor (TcR) resembles that of antibody but consists of only 2 polypeptide chains with a single binding site. Each chain has a constant and variable structural domain. An αβ TcR and a γδ Tcr have been described. Antigen interaction with receptors is noncovalent and reversible. The TcR binding site for antigen (one site per receptor) is also formed by the variable domains of an alpha and beta chain polypeptide. 7
Nomenclature for structural differences between any 2 antibodies. There are 5 immunoglobulin classes based on structural similarity of the H chain C domains. Antigen-Antibody Reactions, Definitions IgM and IgA and Characteristics to Know are secreted as pentamers and dimers • Size of Epitope/antigenic determinant respectively. • specific,noncovalent,reversible binding The J chain protein • affinity vs. avidity promotes • valency, crosslinking, polymerization. agglutination,precipitation • multideterminant antigens, • heterogeneous antibody responses • cross reactions 8
Protective functions of antibody interactions with Soluble antigens antigen. -continued. mixed with equivalent amounts of soluble specific antibody can bind to form lattices (antigen-antibody complexes) large enough to become insoluble and precipitate. Insoluble (particulate) antigens + antibody form clumps (agglutinate). Fig A.9 Protective functions of antibody interactions Protective functions of antibody reactions with antigen. -continued. with antigens. 9
Recommend
More recommend