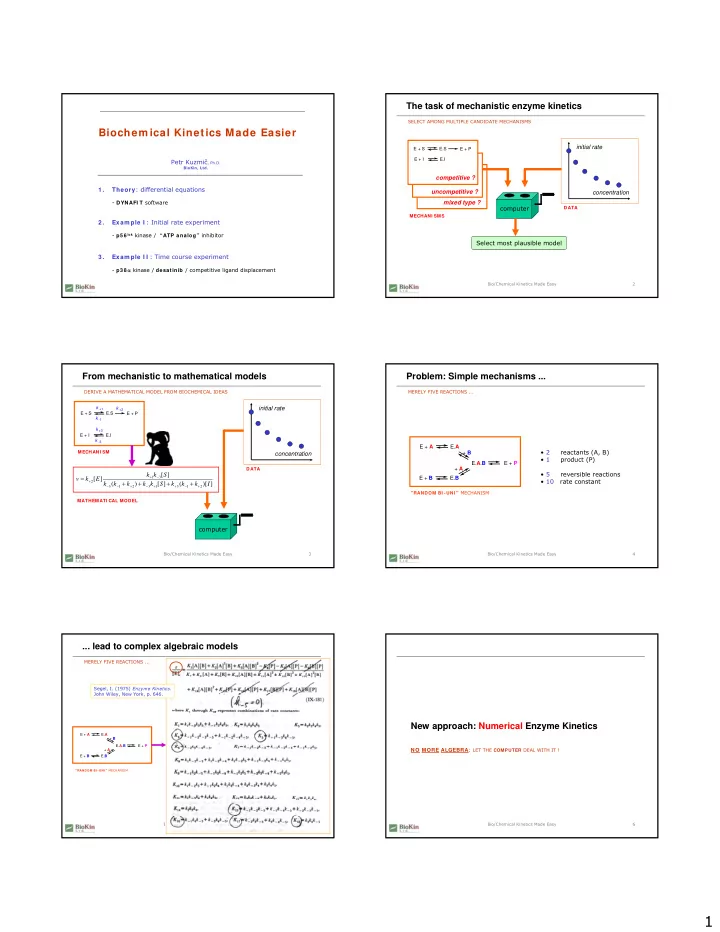

The task of mechanistic enzyme kinetics SELECT AMONG MULTIPLE CANDIDATE MECHANISMS Biochem ical Kinetics Made Easier initial rate E + S E.S E + P E + I E.I Petr Kuzmi č , Ph.D. BioKin, Ltd. competitive ? competitive ? Theory : differential equations 1 . uncompetitive ? concentration - DYNAFI T software mixed type ? computer DATA MECHANI SMS Exam ple I : Initial rate experiment 2 . - p5 6 lck kinase / “ATP analog” inhibitor Select most plausible model Exam ple I I : Time course experiment 3 . - p3 8 α kinase / desatinib / competitive ligand displacement Bio/Chemical Kinetics Made Easy 2 From mechanistic to mathematical models Problem: Simple mechanisms ... DERIVE A MATHEMATICAL MODEL FROM BIOCHEMICAL IDEAS MERELY FIVE REACTIONS ... k +1 initial rate k +2 E + S E.S E + P k -1 k +3 E + I E.I k -3 E + A E. A • 2 reactants (A, B) MECHANI SM + B concentration • 1 product (P) E. A . B E + P DATA + A • 5 reversible reactions k k [ S ] = + − v k [ E ] 1 3 E + B E. B + • 10 rate constant 2 + + + + k ( k k ) k k [ S ] k ( k k )[ I ] − − + − + + − + 3 1 2 3 1 3 1 2 "RANDOM BI -UNI " MECHANISM MATHEMATI CAL MODEL computer Bio/Chemical Kinetics Made Easy 3 Bio/Chemical Kinetics Made Easy 4 ... lead to complex algebraic models MERELY FIVE REACTIONS ... Segel, I. (1975) Enzyme Kinetics . John Wiley, New York, p. 646. New approach: Numerical Enzyme Kinetics E. A E + A + B E. A . B E + P NO MORE ALGEBRA : LET THE COMPUTER DEAL WITH IT ! + A E + B E. B "RANDOM BI - UNI " MECHANISM Bio/Chemical Kinetics Made Easy 5 Bio/Chemical Kinetics Made Easy 6 1
Theoretical foundations: Mass Action Law Theoretical foundations: Mass Conservation Law RATE IS PROPORTIONAL TO CONCENTRATION(S) PRODUCTS ARE FORMED WITH THE SAME RATE AS REACTANTS DISAPPEAR MONOMOLECULAR REACTIONS EXAMPLE A products rate is proportional to [A] A d[ ]/dt d[ ]/dt d[ ]/dt P + Q - A = + P = + Q - d [ A ] / d t = k [ A ] BIMOLECULAR REACTIONS COMPOSITION RULE ADDITIVITY OF TERMS FROM SEPARATE REACTIONS A + B products rate is proportional to [A] × [B] mechanism: - d [ A ] / d t = - d [ B ] / d t = k [ A ] × [B] k 1 d [ B ] / d t = + k 1 [A] - k 2 [B] A B k 2 B C Bio/Chemical Kinetics Made Easy 7 Bio/Chemical Kinetics Made Easy 8 Program DYNAFIT Initial rate kinetics TWO BASIC APPROXIMATIONS REFERENCES 1. Kuzmic P. (1996) Anal. Biochem. 2 3 7 , 260-273. “Program DYNAFIT for the analysis of enzyme kinetic data” 1. Rapid-Equilibrium Approximation 2. Kuzmic P. (2009) Methods in Enzymology , in press “DYNAFIT – A software package for enzymology” assumed very much slow er than k 1 , k 2 k 1 k 3 E + S E. S E + P Ref. [1] – total citations: k 2 5 0 0 FREE TO ACADEMIC USERS 2. Steady-State Approximation 4 0 0 www. biokin .com Mathematical details in BBA – Proteins & Proteom ics , submitted New in 3 0 0 DynaFit 2 0 0 • no assumptions made about relative magnitude of k 1 , k 2 , k 3 1 0 0 0 1 9 9 8 2 0 0 0 2 0 0 2 2 0 0 4 2 0 0 6 2 0 0 8 Bio/Chemical Kinetics Made Easy 9 Bio/Chemical Kinetics Made Easy 10 Initial rate kinetics - Traditional approach Initial rate kinetics in DynaFit DERIVE A MATHEMATICAL MODEL FROM BIOCHEMICAL IDEAS GOOD NEWS: MODEL DERIVATION CAN BE FULLY AUTOMATED! DynaFit input file MATHEMATI CAL MODEL k +1 initial rate k +2 E + S E.S E + P [task] k -1 task = fit 0 = [E] + [E.A] + [E.B] + [E.A.B] – [E] tot data = rates k +3 0 = [A] + [E.A] + [E.A.B] – [A] tot approximation = steady-state 0 = [B] + [E.B] + [E.A.B] – [B] tot E + I E.I 0 = + k 1 [E][A] – k 2 [E.A] – k 3 [E.A][B] + k 4 [E.A.B] k -3 0 = + k 5 [E][B] – k 6 [E.B] – k 7 [E.B][A] + k 8 [E.A.B] [mechanism] 0 = + k 3 [E.A][B] + k 7 [E.B][A] + k 10 [E][P] – ( k 4 +k 8 +k 9 )[E.A.B] derive push MECHANI SM concentration equations button E + A <==> E.A : k1 k2 E.A + B <==> E.A.B : k3 k4 DATA E + B <==> E.B : k5 k6 initial rate k k [ S ] = + − 1 3 v k [ E ] E.B + A <==> E.A.B : k7 k8 + 2 + + + + k ( k k ) k k [ S ] k ( k k )[ I ] − − + − + + − + E.A.B <==> E + P : k9 k10 3 1 2 3 1 3 1 2 [constants] concentration MATHEMATI CAL MODEL ... DATA MECHANI SM computer computer Bio/Chemical Kinetics Made Easy 11 Bio/Chemical Kinetics Made Easy 12 2
Initial rate kinetics in DynaFit vs. traditional method WHICH DO YOU LIKE BETTER? [task] Biochem ical Kinetics Made Easier task = fit data = rates DynaFit applications to protein kinases approximation = steady-state [reaction] A + B --> P [mechanism] Case study #1: I NI TI AL RATES OF ENZYME REACTI ONS E + A E. A + B E + A <==> E.A : k1 k2 E. A . B E + P inhibition constants and kinetic mechanism + A E.A + B <==> E.A.B : k3 k4 E. B E + B E + B <==> E.B : k5 k6 E.B + A <==> E.A.B : k7 k8 E.A.B <==> E + P : k9 k10 [constants] ... [concentrations] ... Bio/Chemical Kinetics Made Easy 13 WIN-61651: Presumably an ATP analog? Lineweaver-Burk plots for WIN-61651 TRADITIONAL STUDY: KINASE INHIBITOR ‘WIN-61651’ IS COMPETITIVE WITH ATP LINEWEAVER-BURK PLOTS AT VARIED [PEPTIDE] AND FIXED [ATP] ARE NONLINEAR Faltynek et al. (1995) J. Enz. Inhib. 9 , 111-122. Faltynek et al. (1995) J. Enz. Inhib. 9 , 111-122. O O O O 15 NH 2 NH 2 N N N N [I] = 0 N N 10 WIN-61651 WIN-61651 N N 1/v N N 5 [I] = 80 μ M 0 0 1 2 1/[RRSRC] Bio/Chemical Kinetics Made Easy 15 Bio/Chemical Kinetics Made Easy 16 Direct plot for WIN-61651: Initial rate vs. [peptide] Adding a substrate inhibition term improves fit MIXED-TYPE INHIBITION MECHANISM: WHICH IS SMALLER, K is or K ii ? GLOBAL NUMERICAL FIT IS BOTH MORE PRECISE AND MORE ACCURATE Faltynek et al. (1995) J. Enz. Inhib. 9 , 111-122. – FI GURE 1B [mechanism] [mechanism] [I] = 0 100 E + S <===> ES 100 E + S <===> ES ES ---> E + P ES ---> E + P ES + S <===> ES2 E + I <===> EI E + I <===> EI ES + I <===> ESI ES + I <===> ESI rate rate 50 50 E.S.S K s2 E E.S E + P E E.S E + P K is K ii K is K ii 0 0 E.I E.S.I E.I E.S.I 0 2000 4000 6000 0 2000 4000 6000 [RRSRC], μ M [RRSRC], μ M Bio/Chemical Kinetics Made Easy 17 Bio/Chemical Kinetics Made Easy 18 3
How do we know which mechanism is "best"? WIN-61651 summary: Comparison of methods COMPARE ANY NUMBER OF MODELS IN A SINGLE RUN WIN-61651 IS A MIXED-TYPE INHIBITOR, NOT COMPETITIVE WITH ATP [task] task = fit | data = rates model = mixed-type ? Faltynek [reaction] | S ---> P [enzyme] | E et al. (1995) DynaFit parameter (mM) [modifiers] | I ... 9100 ± 3700 990 ± 140 K s 1100 ± 450 — K s2 [task] 28 ± 2 1 8 ± 4 competitive: K is task = fit | data = rates 1 4 ± 5 67 ± 18 uncompetitive: K ii model = competitive ? ... residual [task] squares 2 .1 1 9 .5 task = fit | data = rates model = uncompetitive ? Akaike I nform ation Criterion ... Review: Burnham & Anderson (2004) Bio/Chemical Kinetics Made Easy 19 Bio/Chemical Kinetics Made Easy 20 Kinase – Antibody – Tracer – Inhibitor assay A FOUR-COMPONENT MIXTURE Biochem ical Kinetics Made Easier DynaFit applications to protein kinases Case study #2: REACTI ON PROGRESS rate constants for kinase-inhibitor interactions competitive ligand displacement FRET assay 1 2 3 4 Preliminary experimental data: Bryan Marks , Invitrogen (life Technologies) Bio/Chemical Kinetics Made Easy 22 Kinase – Antibody – Tracer – Inhibitor: mechanism Rate constants and receptor-ligand residence time PURPOSE: OBTAIN RATE CONSTANTS FOR INHIBITOR ASSOCIATION & DISSOCIATION IS IT WORTH CHASING AFTER RATE CONSTANTS? Mbalaviele et al. (2009) J. Pharm. Exp. Ther . 3 2 9 , 14-25 “PHA-408 is an ATP competitive inhibitor, which binds I KK-2 tightly with a relatively slow off rate .” Puttini et al. (2008) haem atologica 9 3 , 653-61 E ... e nzyme A ... a ntibody (FRET donor) “The present results suggest a slower off- rate (dissociation T ... t racer (FRET acceptor) rate) of [a novel Abl kinase inhibitor] compared to im atinib as an explanation for the increased cellular activity of the I ... i nhibitor former.” Tummino & Copeland (2008) Biochemistry 4 7 , 5481-92 • four components “... the extent and duration of responses to receptor-ligand • five complexes (3 binary, 2 ternary) • six unique rate constants interactions depend greatly on the tim e period over which the ligand is in residence on its receptor.” Bio/Chemical Kinetics Made Easy 23 Bio/Chemical Kinetics Made Easy 24 4
Recommend
More recommend