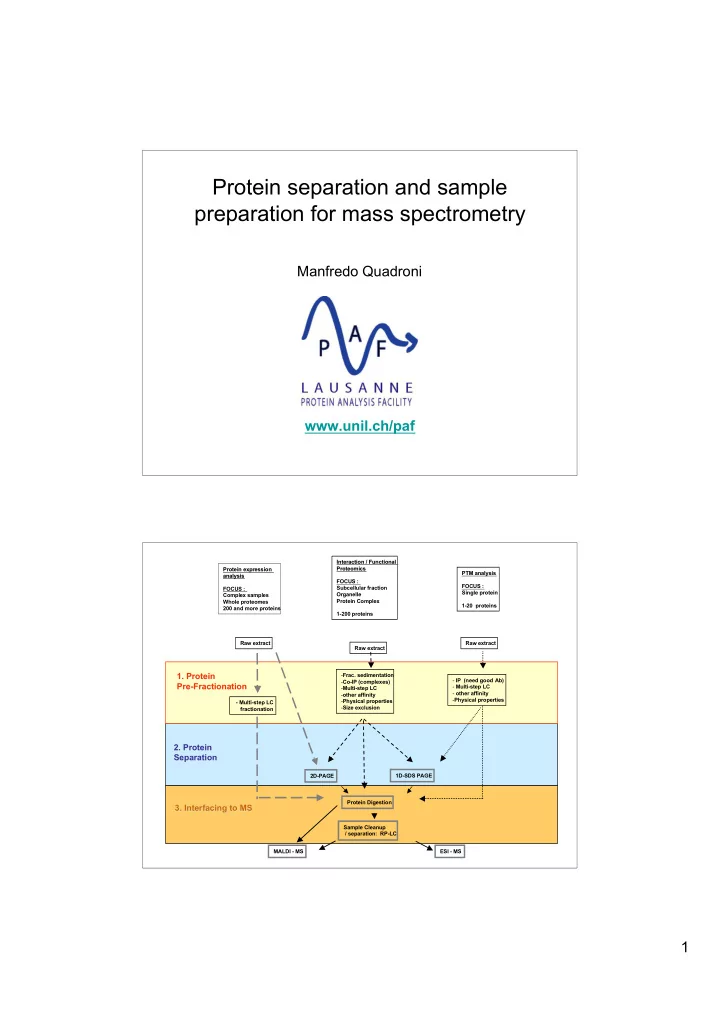

Protein separation and sample preparation for mass spectrometry Manfredo Quadroni www.unil.ch/paf Interaction / Functional Interaction / Functional Protein expression Protein expression Proteomics Proteomics PTM analysis PTM analysis analysis analysis FOCUS : FOCUS : Subcellular fraction FOCUS : Single protein Organelle Complex samples Whole proteomes Protein Complex 1-20 proteins 200 and more proteins 1-200 proteins Raw extract Raw extract Raw extract 1. Protein 1. Protein - Frac. sedimentation - IP (need good Ab) - Co-IP (complexes) Pre-Fractionation Pre-Fractionation - Multi-step LC - Multi-step LC - other affinity - other affinity - Physical properties - Physical properties - Multi-step LC - Size exclusion fractionation 2. Protein 2. Protein Separation Separation 2D-PAGE 1D-SDS PAGE Protein Digestion 3. Interfacing to MS 3. Interfacing to MS Sample Cleanup / separation: RP-LC MALDI - MS ESI - MS 1
Protein separation/fractionation : a revival of old fashioned biochemistry All “classical” methods can be used in all possible combinations : Liquid chromatography : - anion exchange - cation exchange - hydrophobic interaction - hydroxylapatite, heparin,… - batch methods : fractional precipitations - affinity : Immobilised Antibodies, tagged molecules, drugs, peptides, - size exclusion Isoelectric focusing & chromatofocusing Electrophoresis : - SDS-PAGE ( 1D & 2D ) - native (Urea, Blue Native, Triton Acidic Urea) -… Choose methods based on orthogonal separation principles Key points : Choose methods compatible on the basis of sample composition Protein separation/fractionation : a revival of old fashioned biochemistry PRINCIPLE OF RESOLUTION NATIVE OR SCALABILITY SAMPLE RECOVERY IN SEPARATION DENATURING LC : ANION EXCHANGE SURFACE LOW-MEDIUM NATIVE YES BUFFER + SALT CHARGE LC : CATION EXCHANGE SURFACE LOW-MEDIUM NATIVE YES BUFFER + SALT CHARGE LC : HYDROPHOBIC SURFACE LOW-MEDIUM NATIVE YES BUFFER + SOLVENT HYDROPHOBICI TY LC: SIZE EXCLUSION / SIZE LOW-MEDIUM NATIVE YES BUFFER GEL FILTRATION LC:SPECIFIC AFFINITY BINDING N.A. NATIVE* YES BUFFER+ TO MATRIX ADDITIVES* IEF NET CHARGE MEDIUM-HIGH DENATURING MEDIUM BUFFER+AMPHOLYTES+ DENATURANTS 1D-SDS PAGE SIZE HIGH DENATURING LIMITED GEL+SDS+BUFFER 2D SDS PAGE NET CHARGE + VERY HIGH DENATURING LIMITED GEL+SDS+BUFFER SIZE BLUE NATIVE GEL SURFACE MEDIUM NATIVE LIMITED GEL+BUFFER CHARGE 2
After digestion : separation of peptides separation Liquid chromatography : - strong cation exchange (SCX) - strong anion exchange (SAX) separation Separation - reversed-phase (RP) + desalting - immobilised metal affinity (IMAC) P-peptides SCX MS SAX RP Protease IMAC digestion (Trypsin) Interfacing to MS: goals Interfacing to MS: goals Purity separate complex mixtures Cleanliness eliminate molecules that interfere with ionisation/detection 3
Interfacing to MS: “ Interfacing to MS: “Killer Killer” ” substances to eliminate substances to eliminate Most charged or ionisable Most charged or ionisable molecules interfere with the molecules interfere with the ionisation of the analyte (i.e. compete for charges ) and cause ionisation of the analyte (i.e. compete for charges ) and cause signal suppression and/or elevation of the background noise signal suppression and/or elevation of the background noise Salts : Na+, Cl-, Tris, … Chaotropes : Urea, Thiourea, Gu-HCl, Detergents : SDS, NP-40, Triton X, TWEEN,… Polymers : PEG, Ampholytes, .. All non volatile, ionic compounds : glycerol,DMSO,… Interfacing to MS: signal suppression by NP-40 Interfacing to MS: signal suppression by NP-40 44 Da 44 Da np-40/Scans 1-36 +Profile Q3SCAN 44 Da Average of scans 1 to 36 Time=0.55 min trial ppeptides - 1.10.1996 - 12:29 No Title 65,945 Solvent : 44 Da 100 50 % MeOH, 0.5 % Formic acid 44 Da ? % NP-40 44 Da Sample was “cleaned up” on C18 75 -> concentration of detergent rather 44 Da than peptide Relative Intensity (%) 44 Da 44 Da 50 25 0 400 500 600 700 800 900 1000 1100 1200 1300 1400 JW1 Peptide, 4 mg / ml m/z 4
MALDI: MALDI: crystallisation crystallisation is essential is essential Good crystallisation (DHB) No crystallisation due to salt or detergent no signal !!! α -CHCA DHB Liquid samples: removal of Liquid samples: removal of ‘ ‘contaminants contaminants’ ’ before MS before MS Molecule to eliminate Technique Applicable Applicable Further clean-up to proteins to peptides by SDS HILIC + ++ Reversed-phase SCX Non ionic detergents (NP- SCX (+) + Reversed-phase 40, TWEEN, Triton x100) Zwitterionic detergents SCX or Reversed- ++ ++ Reversed-phase (CHAPS) phase C18, C8,C4 PEGs SCX (+) + Reversed-phase Salts Reversed-phase C18, + +++ - C8,C4 Chaotropes (Urea) Reversed-phase C18, + +++ - C8,C4 Buffers Reversed-phase C18, + +++ - C8,C4 Ampholytes ? - - - Many SDS-PAGE ++++ - Reversed-phase SCX : strong cation exchange ; HILIC : hydrophobic interaction chromatography 5
Liquid samples: removal of ‘ ‘contaminants contaminants’ ’ before MS before MS Liquid samples: removal of Better than all the above Better than all the above Design protocols to avoid use of some of (these) contaminants Design protocols to avoid use of some of (these) contaminants not always possible but worth a try not always possible but worth a try • • Reduce detergents Reduce detergents • Substitute polymeric detergents (NP-40, Triton,TWEEN) with • Substitute polymeric detergents (NP-40, Triton,TWEEN) with others with low molecular weight and defined molecular composition: others with low molecular weight and defined molecular composition: CHAPS, CHAPS, Octyl Octyl Glucoside Glucoside • Use volatile buffers ( ammonium acetate, ammonium bicarbonate) Use volatile buffers ( ammonium acetate, ammonium bicarbonate) • in final steps in final steps • • Include desalting at the end of purification schemes Include desalting at the end of purification schemes Interfacing biochemistry to MS Interfacing biochemistry to MS • SDS-PAGE : 1D (or 2D) are a nearly universal interface between sample preparation and MS (for protein ID or PTM studies) • LC-based approaches : more care is required in planning upstream experiments 6
Why is SDS-PAGE such a good preparation method? Why is SDS-PAGE such a good preparation method? • Ideal interface to biology • Analytical and micropreparative • Robust • Solid phase chemistry of proteins • Easy, low-tech • Removal of contaminants : – At the loading point – After migration during fix / staining steps • protein digestion in gel: non quantitative Disadvantages • peptide sequence recovery: usually incomplete • whole protein recovery: poor In-gel digestion: solid phase chemistry of proteins In-gel digestion: solid phase chemistry of proteins 7
From 1D- and 2D-PAGE to MS: sample preparation From 1D- and 2D-PAGE to MS: sample preparation Protease Peptide digestion extraction (Trypsin) Desalting on rp column MS Sequence Best matching sequence database m/z • Spot cutting and sample processing can be automated Interfacing to MS: conclusions Interfacing to MS: conclusions • SDS-PAGE and reversed-phase LC are the main final interfaces between biology and Mass Spectrometry; these techniques can be used both to clean up and separate proteins before MS • Many compounds commonly used in protein purification are incompatible with MS • Hence: every new experiment should be planned from the beginning to optimise the preparation of the sample for MS – Efficiency – Sensitivity – Fractionation of complex mixtures 8
Introduction to MALDI and Electrospray mass spectrometry Manfredo Quadroni www.unil.ch/paf Mass distribution and mass measurements Isotope Atomic mass % Abundance Element Mr(mono.) Mr(avg.) 1H 1.0078 99.985 H 1.0078 1.0080 2H 2.0141 0.015 ----------------------------------------------------------------------------------------------------------------------------------- 12C 12.0000 98.93 C 12.0000 12.0107 13C 13.0034 1.07 ------------------------------------------------------------------------------------------------------------------------------------ 35Cl 34.9689 75.78 Cl 34.9689 35.4525 37Cl 24.22 24.22 Sequence Composition MW(mono) MW(ave) m/z (mono) m/z(ave) ------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------ VATVSLPR C37.H68.N11.O11 841.5022 841.999 842.510 843.014 LGEHNIDVLEGNEQFINAAK C96.H152.N27.O33 2210.097 2211.397 2211.105 2211.105 m/z = (m + (mA * z )) / z -> m/z (mono) of VATVSLPR 1+ : ( 841.502 + 1.008 ) /1 = 842.510 -> m/z (mono) of LGE…AAK 3+ : ( 2210.097 + (3 x 1.008 )) /3 = 737.707 9
Singly charged ions as an example Mono. Mono. % Intensity % Intensity 842.502 100 2212.120 100 Avg. 90 Avg. 90 80 80 2213.137 70 2211.097 70 60 60 50 843.485 50 40 2214.149 40 30 30 844.480 20 20 10 10 0 0 0 0 2209.0 2210.6 2212.2 2213.8 2215.4 2217.0 839.0 840.6 842.2 843.8 845.4 847.0 Mass (m/z) Mass (m/z) Theoretical MS : the effect of resolution 100 90 Theoretical distribution 80 Calculated with resolution 1000 70 60 Calculated with resolution 10000 50 40 30 20 10 0 842 844 846 848 850 10
Recommend
More recommend