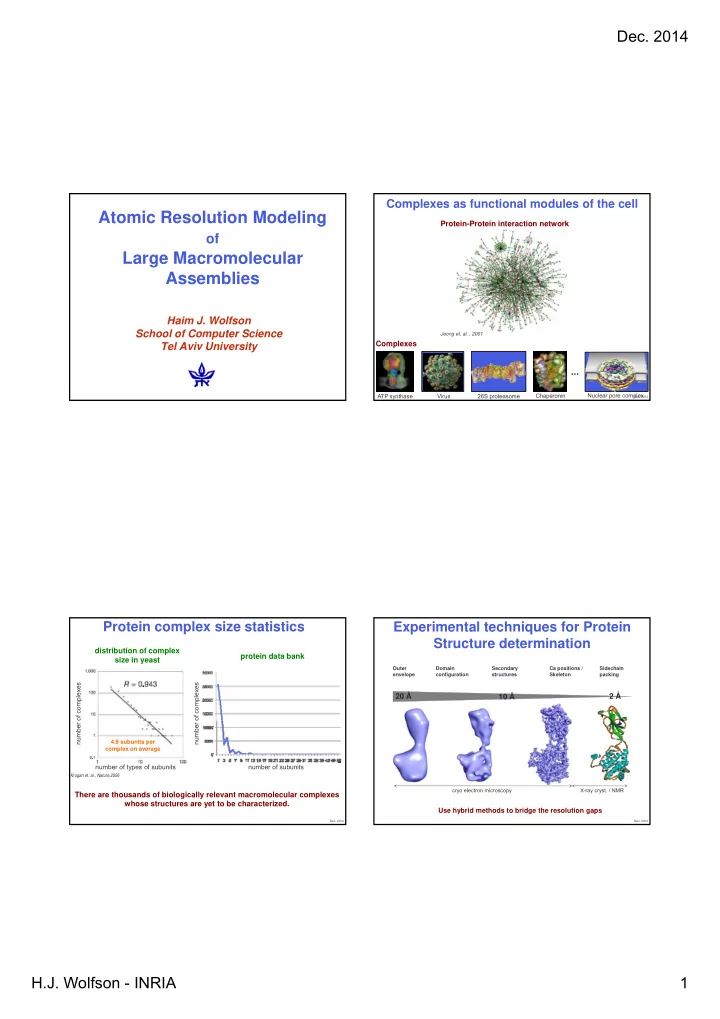

Dec. 2014 Complexes as functional modules of the cell Atomic Resolution Modeling Protein-Protein interaction network of Large Macromolecular Assemblies Haim J. Wolfson School of Computer Science Jeong et. al. , 2001 Complexes Tel Aviv University ... ATP synthase Virus 26S proteasome Chaperonin Nuclear pore complex Dec. 2014 Protein complex size statistics Experimental techniques for Protein Structure determination distribution of complex protein data bank size in yeast Outer Domain Secondary Ca positions / Sidechain envelope configuration structures Skeleton packing of complexes of complexes 20 Å 10 Å 2 Å number number 4.9 subunits per complex on average number of types of subunits number of subunits Krogan et. al., Nature,2006 cryo electron microscopy X-ray cryst. / NMR There are thousands of biologically relevant macromolecular complexes whose structures are yet to be characterized. Use hybrid methods to bridge the resolution gaps Dec. 2014 Dec. 2014 H.J. Wolfson - INRIA 1
Dec. 2014 Analogy Puzzle Assembly in Computer Vision and Multi-molecular assembly is analogous to the solution of 3D Robotics puzzles –a classical spatial Pattern Discovery task. High resolution data Low resolution data Circa 1 9 8 6 Dec. 2014 Dec. 2014 SPECI AL FREQUENT CASE: Additional Low Resolution Data Sources • FRET Structure Prediction of ( cyclically) • Existence of di-sulfide bonds Sym m etric Multi-Molecular Assem blies • MasSpec (e.g.distance constraints by chemical cross linking). • SAXS • SAXS • Interaction Data (Y2H, gene fusion, similarity with known complexes, etc.) • and more… D. Schneidman-Duhovny et al., Proteins , 60, 217--223, (2005) . D. Schneidman-Duhovny et al., NAR 33 (web server issue), W363—W367, (2005). Dec. 2014 H.J. Wolfson - INRIA 2
Dec. 2014 Cyclic Sym m etry Cyclic Sym m etry Exploiting the Symmetry Constraints • Cyclic symmetry is defined by rotation of a single unit around an axis . • A trivial “naïve” approach – perform “regular” • The angle is determined by a number of units n . multimolecular docking and discard non-symmetric solutions. side view top view symmetry axis • A more sophisticated approach – use the symmetry constraints as an integral part of the algorithm to reduce complexity and improve accuracy complexity and improve accuracy. • Observation – if point A in the protein is matched after the symmetry rotation to point B, one can detect a plane to which the symmetry axis is perpendicular and its location is restricted to a known circle in that plane. Dec. 2014 Dec. 2014 Geometric Analysis The Algorithm l AB l 0 • For each pair of matching interest points A l and B – Calculate C AB α d AB A • For δ = 0 to 360- ∆ step ∆ C • Calculate l C δ d A r tan α • Calculate T l α 2 2 r r 2 2 • If T is valid add T to the candidate transformation list d B r cot B • Cluster transformations 2 2 • Calculate the score for transformations, r is a function of d and α only l is tangent to a circle C of radius r which are cluster representatives T( A ) B which is centered at (A+B)/2 and lies on a plane orthogonal to AB where T T' R (T' is a translati on and R is a rotation) R Dec. 2014 Dec. 2014 H.J. Wolfson - INRIA 3
Dec. 2014 CAPRI Target 1 0 : Chaperon: 2.5 Å RMSD prediction for the homo-heptamer. 9 .0 Å RMSD prediction for the hom o-trim er of a viral coat protein Our Prediction Crystal Structure Dec. 2014 Dec. 2014 Exploit Low Resolution Info – EM, SAXS, FRET etc. Structural models of the subunits Previous Work Low/Medium resolution EM density map at atomic level Early work : Fitting of atomic structures to the density map by cross-correllation. In essence – structural alignment at different resolutions. Recent work : Hybrid Methods. Dec. 2014 Dec. 2014 H.J. Wolfson - INRIA 4
Dec. 2014 Publications MultiFit • W. Wriggers, R.A. Milligan, J.A. McCammon, Situs: a package for docking crystal structures into low resolution maps for electron microscopy, J. Struct. Biol. 125, (1999), 185—195. • Z. Yang, K. Lasker, D. Schneidman-Duhovny, B. Webb, C.C. Huang, E.F. Petersen, T. D. Goddard, E.C. Meng, A. C.C. Huang, E.F. Petersen, T. D. Goddard, E.C. Meng, A. Sali, T.E. Ferrin, UCSF Chimera MODELLER, and IMP: An integrated modeling system, J. Struct. Biol. 179, (2011), 269—278. Find the placements ( translation and orientation) of • E. Karaca, A.S.J. Melquiond, S.J. deVries, P.L. Kastritis atomic components in the density map of their and A.M.J.J. Bonvin, Building Macromolecular association. Assemblies by Information-driven Docking : Introducing Lasker, Topf, Sali, Wolfson, JMB 2009 the HADDOCK MultiBody docking server, Mol. Cel. Proteomics 9, (2010), 1784—1794. Lasker, Sali, Wolfson, Proteins 2010 Dec. 2014 Dec. 2014 MultiFit: A geometric view MultiFit - Example of a Task :Assemble the Input: Arp2/3 structure Number of protein subunits and their structural models Low resolution density map of the entire assembly Goal: Determine the assembly configuration optimizing Goal: Determine the assembly configuration simulated at 20 Å resolution optimizing Structural component %seq id C RMSD docking docking S = Rpb1 40 5.1 resolution Rpb2 48 2.5 alignment ARPC1 16 6.1 Geometric Envelope COMPONENT STRUCTURE – Fitting score complementarity penetration ARPC2 29 21.4 ARPC3 99 0.4 Structural accuracy OUTPUT of HOMOLOGY MODELING ARPC4 29 14.3 Find the placements ( translation and orientation) of atomic components ARPC5 94 5.5 in the density map that minimizes the scoring function Dec. 2014 Dec. 2014 H.J. Wolfson - INRIA 5
Dec. 2014 Few representative reasons for the Focus the subunit placement search difficulty of multiple fitting around anchor points • Scoring • Cross-correlation measure alone is not always sufficient • anchor graph: a low-resolution description of the assembly. to place a component in the map. • nodes: points in 3D that approximate the centroid positions of the • Cross-correlation score does not check for geometric assembly components. complementary between interacting components. • edges: between nodes that are close in space. • Docking alone is problematic, since the accuracy of docking • The anchor graph was constructed using a Gaussian Mixture methods depends on the accuracy of the individual atomic methods depends on the accuracy of the individual atomic Model segmentation of the density map. Model segmentation of the density map structures Pair of components Pairwise docking rank Solution: use a scoring function that considers fitting ARP3/ARPC2 12185 ARP3/ARPC3 854 and geometric complementarity simultaneously ARP3/ARPC4 5888 ARPC1/ARP2 4663 • Optimization ARPC1/ARPC5 5504 • Sequential fitting or sequential pairwise docking may not result in the right configuration in the general case. • Enumerating all possible configurations of components of large The anchor graph Sampling of subunit centroids at anchor graph pts assemblies is too expensive Dec. 2014 Dec. 2014 Reduce the multiple fitting problem to optimization Graphical Models of a subunit location and orientation graph 1. Represent the scoring function as a weighted graph. • Use a belief propagation type algorithm to detect the optimal solution. S = • Apply the algorithm both in the placement stage and Geometric Fitting score Envelope orientation refinement stages. complementarity penetration • Utilise the Junction Graph structure Utilise the Junction Graph structure. Fitting score & Envelope penetration Complexity ???!!! Geometric complementarity Dec. 2014 Dec. 2014 H.J. Wolfson - INRIA 6
Dec. 2014 Reducing the complexity of the scoring graph DOMINO: Optimize large systems by optimization of smaller tractable sub ‐ systems Given a mapping of components to the nodes of the min P ( x 1 ,...., x N ) anchor graph, we can eliminate interaction terms between nodes that are far in space. sequential message passing / Belief propagation / dynamic programming 1 2 i j N Subset minimization S b i i i i Subset minimization S b i i i i Fitting score & Envelope Passing messages on a (junction) tree penetration j Geometric 1 2 complementarity i N S = i Geometric Fitting score Envelope complementarity penetration Dec. 2014 Dec. 2014 Refinement by docking partner MultiFit / DOMINO enrichment Lasker, Topf, Sali, and Wolfson. J. Mol. Biol . 388, 180-194, For each of the top 50 configuration solutions 2009. Map segmented into anchor graph Input: components, map Discretize Sample the placements of each map component by constrained rigid pairwise docking (PatchDock). Iterate over all mappings of components to anchor nodes via branch-and-bound Gather subset Gather subset solutions into the Decompose set solutions into best global of components best possible global solutions solutions. Output: component “Decoupled” subsets of configuration, to be Scoring function as a graph. components. refined. Component fits in vicinity of Sample subsets PatchDock: Duhovny-Schneidman, Nussinov, Wolfson , WABI 2002. their anchor nodes. “independently”. Dec. 2014 Dec. 2014 H.J. Wolfson - INRIA 7
Recommend
More recommend