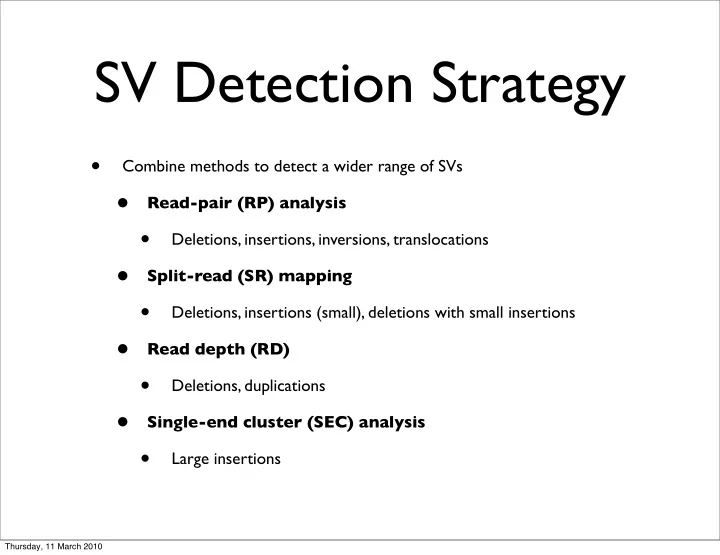

SV Detection Strategy • Combine methods to detect a wider range of SVs • Read-pair (RP) analysis • Deletions, insertions, inversions, translocations • Split-read (SR) mapping • Deletions, insertions (small), deletions with small insertions • Read depth (RD) • Deletions, duplications • Single-end cluster (SEC) analysis • Large insertions Thursday, 11 March 2010
*.bam file SE Cluster CND BreakDancer Pindel Single-end mapped clusters; HMM ReadDepth Read Pairs Split reads large insertions Filter calls by posterior probabilities Filter calls by score Filter calls by score Filter calls by # supporting reads Exclude calls near gaps, cen./tel. Exclude calls near gaps, cen./tel. Exclude calls near gaps, cen./tel. Exclude calls near gaps, cen./tel. Min. Loss = 1kb, Min. Gain= 2kb Separate small (<100bp) and large SVs • Merge overlapping SVs of the same type • Create tab-delimited (BED) ‘merged’ SV list • Run local assemblies • Align contigs to Reference Summary Stats • Total deletions, Overlap SVs with: insertions, etc. Genes/Exons • Number of SVs QTL regions affecting exons Other regions of interest Parse contig alignments • Affected genes in QTL • Refine Coordinates • Rank Calls based on alignment evidence Analysis pipeline for calling SVs in Mouse genomes Thursday, 11 March 2010
Breakdancer K. Chen et al ., Nature Methods (2009) • Performs read-pair analysis • Accepts Maq map files or bam files • Handles multiple library insert sizes • Provides a confidence score and number of supporting reads Thursday, 11 March 2010
Read Pair Analysis 200bp insert 200bp insert Deletion Insertion ref ref Inversion Translocation ref ref A B Thursday, 11 March 2010
Read pairs displayed in LookSeq. Deletions are easily spotted; read pairs are mapped further apart than expected Thursday, 11 March 2010
Mate pairs align in the same orientation Dip in coverage at the breakpoints Inversion on chr 4 Thursday, 11 March 2010
Pindel K. Ye et al ., Bioinformatics (2009) • Maq can’t align reads at breakpoints of larger SVs • Often one read in a pair is mapped, other is unmapped • Pindel uses mapped partner to localise unmapped reads, and uses a split-read approach to align the read (del:1bp - 100kb, ins:1bp - <100bp) 200bp insert 200bp insert Insertion Deletion ref ref Thursday, 11 March 2010
CND J. Simpson et al ., Bioinformatics (2010) • Copy number detection from mapped read depth using a Hidden Markov Model (HMM) • Resolution: 1kb • Uses bam files and samtools pileup • Repeat regions are skipped • GC correction included to improve calls Thursday, 11 March 2010
GC content of mapped reads from individual lanes Thursday, 11 March 2010
Total Mb included in regions of copy number gain or loss, with and without GC correction Corrected depth for each 1 kb window: m d = d i . Gain, no GC correction m GC Gain, GC correction where d i is the mean depth per base of the Loss, no GC correction i th window, m GC is the median depth of all Loss, GC correction windows with the same G+C percentage as the i th window, and m is the median depth of all windows (revised from Yoon et al., 2009) Thursday, 11 March 2010
SE Clusters • Identifies candidate insertion sites by finding clusters of single-end mapped reads (one end mapped, other end unmapped) Unmapped reads inserted sequence ref Thursday, 11 March 2010
*.bam file SE Cluster CND BreakDancer Pindel Single-end mapped clusters; HMM ReadDepth Read Pairs Split reads large insertions Filter calls by posterior probabilities Filter calls by score Filter calls by score Filter calls by # supporting reads Exclude calls near gaps, cen./tel. Exclude calls near gaps, cen./tel. Exclude calls near gaps, cen./tel. Exclude calls near gaps, cen./tel. Min. Loss = 1kb, Min. Gain= 2kb Separate small (<100bp) and large SVs • Merge overlapping SVs of the same type • Create tab-delimited (BED) ‘merged’ SV list ‘Merged’ set • Run local assemblies • Align contigs to Reference Summary Stats • Total deletions, Overlap SVs with: insertions, etc. Genes/Exons • Number of SVs QTL regions affecting exons Other regions of interest Parse contig alignments • Affected genes in ‘Refined’ set QTL • Refine Coordinates • Rank Calls based on alignment evidence Analysis pipeline for calling SVs in Mouse genomes Thursday, 11 March 2010
Filtering and merging • SV are filtered by score or number of supporting reads; HMM calls filtered by posterior probabilities • Remove SVs near reference sequence gaps, and1Mb from centromere or telomere • Reference mm9: 562 gaps (~96 Mb total) • Take the union of calls from all methods to create a ‘Merged’ set Thursday, 11 March 2010
Applying several methods to identify SVs Deletions NOD strain 13,442 4377 544 BreakDancer Pindel (min 100bp) Insertions NOD strain 21 11359 934 Pindel BreakDancer 3 (all calls are >100bp) 11 275 Overlap between methods is 3957 small; Using only a single method will not provide a SECluster complete list of SVs Thursday, 11 March 2010
Local assembly and breakpoint refinement • Local assemblies (Velvet) are performed for each SV in ‘merged’ set, except CND calls • Two assemblies for each, with scaffolding and no scaffolding • Contigs are aligned (Exonerate) to reference, alignments parsed to find breakpoint(s); SVs are ranked and breakpoint coordinates adjusted: • Expected SV is found within range: rank 1 (all rank 1 are in the ‘refined’ set ) • Expected SV not found, the alternate SV is recorded: rank 1 • Expected SV not found, there are gaps in contig coverage: rank 2 (inconclusive) • No breakpoints are found, no large gaps in contig coverage: rank 3 Thursday, 11 March 2010
Complications • Local assembly from mapped reads • Unmapped mates are included, but large insertions can’t be completely assembled (eg: both reads are inside the insertion, they may be ‘unmapped’) • Reads are not aligned near breakpoints if too many SNPs/indels are present (scaffolding helps sometimes) • Automated parsing of alignments: • Repetitive sequence or microhomology at or flanking breakpoints • Variants and small indels near breakpoints Thursday, 11 March 2010
Examples SV calls confirmed by local assembly Thursday, 11 March 2010
Inversion Insertion Insertion (zoomed in) LookSeq view of Chr 4 inversion + insertion Thursday, 11 March 2010
Chr 4 inversion - Local assembly - NODE_1 and NODE_2 contigs Ins inversion contain breakpoints UCSC browser view of aligned contigs Thursday, 11 March 2010
Chr 6 LTR deletion Mate pairs span an LTR in the reference genome Orange reads have mapping quality 0 (they map to more than one location). Thursday, 11 March 2010
Chr 6 LTR deletion Contig spans the breakpoints - The repetitive reads aligned to this region by Maq are also assembled - Local assembly - No scaffolding UCSC browser deletion view of aligned contigs Thursday, 11 March 2010
Chr 13 insertion Clusters of single-end mapped reads (green) Dip in coverage Thursday, 11 March 2010
Chr 13 insertion - Local assembly - Scaffolding - Contig alignments are cut off near the expected breakpoint location - Part of insertion is reconstructed with scaffolding UCSC browser view of aligned contigs Thursday, 11 March 2010
Chr 13 insertion Contig 1 BLAST: Both Celera assembly hit contigs align full- Reference assembly hit length to Mouse N’s from scaffolding Celera assembly* Contig 2 Reference assembly hit Celera assembly hit *The Celera assembly is a mixed-strain assembly of 129X1/SvJ, DBA/2J, and A/J (Released in 2001) Thursday, 11 March 2010
Chr 17 deletion Thursday, 11 March 2010
Chr 17 deletion - Local assembly - No scaffolding - No contig coverage in the deleted region, but no contig crosses the breakpoint UCSC browser view of aligned contigs Thursday, 11 March 2010
Chr 17 deletion - Scaffolding allows contigs to join and provides evidence for the deletion UCSC browser view of aligned contigs Thursday, 11 March 2010
Analysis of 17 inbred mouse Genomes Thursday, 11 March 2010
Local assembly of deletions in Mouse (predicted size >=100bp) 90000 Confirm(>=75bp) Confirm(<75bp) Complex Gap NoMatch NoAssem 67500 Deletions are easiest to predict and find exact breakpoints for using local assembly 45000 22500 0 D J J i J h i J J 2 J N J i J 5 O E E E O m e _ P _ _ P _ _ 2 S _ _ 6 _ Z H P _ A R 9 c A _ 9 N I T s _ B N v L K B K 2 B A 2 _ S u L S S H L B A W t C A 1 B 1 _ e A W 3 7 D 1 C P r C p B 5 S S C 9 2 1 - Confirm(<75): local assembly identified a deletion, but less than 75bp - Complex: event involving a deletion and another event - Gap: not enough contig coverage (inconclusive) - NoMatch: No deletion found, but another SV type may be found - NoAssem: predicted deletion is in very high coverage region Thursday, 11 March 2010
Recommend
More recommend