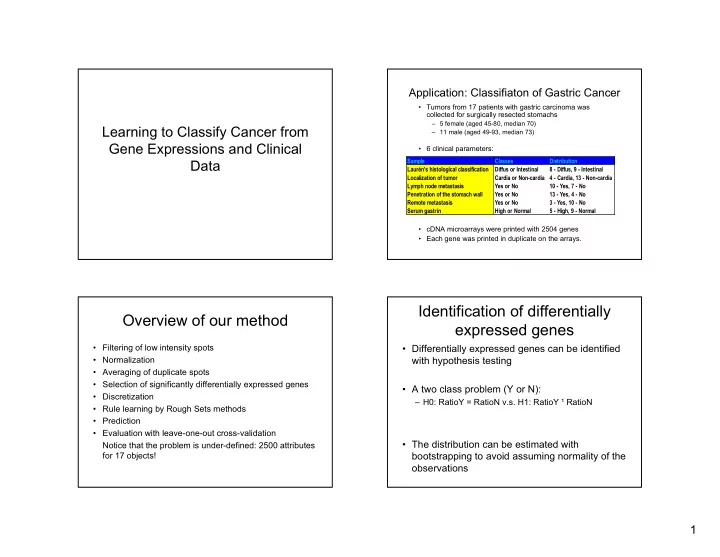

Application: Classifiaton of Gastric Cancer • Tumors from 17 patients with gastric carcinoma was collected for surgically resected stomachs – 5 female (aged 45-80, median 70) Learning to Classify Cancer from – 11 male (aged 49-93, median 73) Gene Expressions and Clinical • 6 clinical parameters: Data Sample Classes Distribution Laurén's histological classification Diffus or Intestinal 8 - Diffus, 9 - Intestinal Localization of tumor Cardia or Non-cardia 4 - Cardia, 13 - Non-cardia Lymph node metastasis Yes or No 10 - Yes, 7 - No Penetration of the stomach wall Yes or No 13 - Yes, 4 - No Remote metastasis Yes or No 3 - Yes, 10 - No Serum gastrin High or Normal 5 - High, 9 - Normal • cDNA microarrays were printed with 2504 genes • Each gene was printed in duplicate on the arrays. Identification of differentially Overview of our method expressed genes • Filtering of low intensity spots • Differentially expressed genes can be identified • Normalization with hypothesis testing • Averaging of duplicate spots • Selection of significantly differentially expressed genes • A two class problem (Y or N): • Discretization – H0: RatioY = RatioN v.s. H1: RatioY ¹ RatioN • Rule learning by Rough Sets methods • Prediction • Evaluation with leave-one-out cross-validation • The distribution can be estimated with Notice that the problem is under-defined: 2500 attributes for 17 objects! bootstrapping to avoid assuming normality of the observations 1
Gene selection Classification Gene selection with bootstraping for Lymph Decision system: PMAIP1 ENPEP GTF3C1 CACNB1 HPN DKFZP434J1813 TGFB3 MGC8471 ... Class Node Metastasis: [*, 0.036) [*, -0.046) [*, -0.226) [-0.136, 0.290) [*, -0.288) [*, -0.044) [*, -0.152) [-0.016, 0.318) ... Y [0.036, 0.440) [0.380, *) [0.026, *) [0.290, *) [0.064, *) [0.292, *) [0.108, *) [0.318, *) ... Y [0.440, *) [0.380, *) [0.026, *) [-0.136, 0.290) [-0.288, 0.064) [0.292, *) [-0.152, 0.108) [0.318, *) ... Y H 0 : Ratio LNM = Ratio Not LNM v.s. H 1 : Ratio LNM ≠ Ratio Not LNM [*, 0.036) [*, -0.046) [*, -0.226) [*, -0.136) [*, -0.288) [*, -0.044) [*, -0.152) [*, -0.016) ... N [0.440, *) [0.380, *) [*, -0.226) [0.290, *) [-0.288, 0.064) [-0.044, 0.292) [0.108, *) [0.318, *) ... Y [*, 0.036) [-0.046, 0.380) [-0.226, 0.026) [-0.136, 0.290) [0.064, *) [-0.044, 0.292) [0.108, *) [-0.016, 0.318) ... Y Cluster(Hs.) Name Symbol Mean P-val boot-t [0.036, 0.440) [*, -0.046) [*, -0.226) [-0.136, 0.290) [*, -0.288) [*, -0.044) [-0.152, 0.108) [-0.016, 0.318) ... N Hs.291 glutamyl aminopeptidase (aminopeptidase A) ENPEP 0.727 0.142 0 [0.440, *) [0.380, *) [-0.226, 0.026) [0.290, *) [0.064, *) [0.292, *) [0.108, *) [0.318, *) ... Y Hs.823 hepsin (transmembrane protease, serine 1) HPN 0.542 0.1238 0 [0.036, 0.440) [*, -0.046) Undefined [*, -0.136) Undefined [*, -0.044) [*, -0.152) [*, -0.016) ... N Hs.74861 activated RNA polymerase II transcription cofactor 4 PC4 0.839 0.1602 0 Undefined [-0.046, 0.380) Undefined Undefined Undefined Undefined [*, -0.152) Undefined ... N Hs.60478 ESTs, Moderately similar to S47073 finger protein HZF2 <Hs.60478> 0.6935 0.1414 0.001 [*, 0.036) [*, -0.046) [-0.226, 0.026) [*, -0.136) [-0.288, 0.064) [-0.044, 0.292) [0.108, *) [*, -0.016) ... N Hs.284266 hypothetical protein MGC8471 MGC8471 0.4589 0.1013 0.001 [0.440, *) [-0.046, 0.380) [0.026, *) [0.290, *) [-0.288, 0.064) [0.292, *) [*, -0.152) [0.318, *) ... Y Hs.96 phorbol-12-myristate-13-acetate-induced protein 1 PMAIP1 0.7117 0.1662 0.002 [0.036, 0.440) [-0.046, 0.380) [*, -0.226) [*, -0.136) [*, -0.288) [-0.044, 0.292) [-0.152, 0.108) [*, -0.016) ... N Hs.2025 transforming growth factor, beta 3 TGFB3 0.5842 0.1585 0.002 [0.036, 0.440) [-0.046, 0.380) [-0.226, 0.026) [-0.136, 0.290) [-0.288, 0.064) [-0.044, 0.292) [-0.152, 0.108) [-0.016, 0.318) ... Y Hs.83469 nuclear factor (erythroid-derived 2)-like 1 NFE2L1 0.8419 0.1812 0.002 [0.440, *) [0.380, *) [0.026, *) [0.290, *) [0.064, *) [0.292, *) [-0.152, 0.108) [-0.016, 0.318) ... Y Hs.181046 dual specificity phosphatase 3 (vaccinia virus phosphatDUSP3 0.4843 0.0943 0.002 [0.036, 0.440) [-0.046, 0.380) [0.026, *) [-0.136, 0.290) [0.064, *) [-0.044, 0.292) [-0.152, 0.108) [-0.016, 0.318) ... Y Hs.331 general transcription factor IIIC, polypeptide 1 GTF3C1 0.2834 0.0864 0.003 [*, 0.036) [*, -0.046) [-0.226, 0.026) [*, -0.136) [*, -0.288) [*, -0.044) [*, -0.152) [*, -0.016) ... N Hs.635 calcium channel, voltage-dependent, beta 1 subunit CACNB1 0.5364 0.122 0.003 Hs.1066 small nuclear ribonucleoprotein polypeptide E SNRPE 0.4673 0.1015 0.003 Hs.1098 DKFZp434J1813 protein DKFZP434J1813 0.5033 0.126 0.003 Decision rules: PMAIP1([*, 0.036)) AND PC4([-0.716, -0.073)) => Class(Y) Hs.104481 Nck, Ash and phospholipase C binding protein NAP4 0.6001 0.1276 0.003 PMAIP1([*, 0.036)) AND PC4([*, -0.716)) => Class(N) Hs.118825 mitogen-activated protein kinase kinase 6 MAP2K6 0.2853 0.0754 0.003 PMAIP1([0.036, 0.440)) AND PC4([*, -0.716)) => Class(N) Hs.161 cadherin 2, type 1, N-cadherin (neuronal) CDH2 0.3771 0.1175 0.004 TGFB3([*, -0.152)) AND MGC8471([-0.016, 0.318)) => Class(Y) Hs.13063 transcription factor CA150 CA150 0.655 0.1831 0.004 TGFB3([0.108, *)) AND MGC8471([-0.016, 0.318)) => Class(Y) Hs.124029 inositol polyphosphate-5-phosphatase, 40kD INPP5A 0.9106 0.2486 0.004 Hs.170980 KIAA0948 protein KIAA0948 0.683 0.1597 0.004 CLCN6([-0.209, 0.141)) AND MGC8471([-0.016, 0.318)) => Class(Y) Hs.211614 chloride channel 6 CLCN6 0.4511 0.114 0.004 CLCN6([*, -0.209)) AND MGC8471([-0.016, 0.318)) => Class(N) Validation from biomedical Prediction Performance literature Sample Reducer Discretation Max Genes Sig. lev. Accuracy Sens. Spec. AUC Laurén's histological classification Dynamic Freq.bin (4) 10 0.01 16/17=0.941 1 0.86 0.93 Known connection to the Localization of tumor Dynamic Entropy 20 0.01 17/17=1 1 1 1 parameter in Known Connection to Unkown Lymph node metastasis Dynamic Freq.bin (3) 20 0.01 14/17=0.824 0.7 1 0.9 Sample gastric cancer other cancer gastric cancer other cancer Connection Penetration of the stomach wall Holte 1r Entropy 20 0.01 16/17=0.941 1 0.75 0.85 Laurén's histological classification 1 2 Remote metastasis Holte 1r Entropy 40 0.1 13/13=1 1 1 1 Localization of tumor 2 3 22 Serum gastrin Genetic Entropy 10 0.05 11/14=0.786 0.9 0.6 0.66 Lymph node metastasis 1 2 1 26 Penetration of the stomach wall 4 1 1 17 Remote metastasis 3 2 47 Serum gastrin 1 1 18 Sample Rules No. (avg) Rules No. (Range) Total no. of genes in all classifiers Laurén's histological classification 24.1 10-67 17 Localization of tumor 238.1 200-311 72 Lymph node metastasis 388.1 222-523 73 Penetration of the stomach wall 109.6 28-280 75 Remote metastasis 425.1 305-468 161 Serum gastrin 47.9 18-72 42 2
Genes occurring in the classifier The classifiers at the best for lymph node metastasis filtering level Symbol Name Function No classifiers Highest level LNM Not LNM LOC51058 hypothetical protein unknown 17 x ISG15 interferon-stimulated protein, 15 kDa signal transduction 17 x Homo sapiens cDNA FLJ14959 fis, clone PLACE4000156unknown 16 x Homo sapiens, clone IMAGE:3948563 unknown 16 x DKFZP434J1813 DKFZp434J1813 protein unknown 16 x CACNB1 calcium channel, voltage-dependent, beta 1 subunit muscle contraction 15 x Homo sapiens, clone MGC:2492, mRNA, complete cds unknown 15 x NAP4 Nck, Ash and phospholipase C binding protein signal transduction 15 x cell division/prot PPP1CC protein phosphatase 1, catalytic subunit, gamma isoform synt 14 x ESTs, Mod similar to JC5238 galactosylceramide-like pro unknown 13 x HAT1 histone acetyltransferase 1 DNA packaging 13 x MGC8471 hypothetical protein MGC8471 unknown 13 x SEC4L GTP-binding prot homo to Sacc cerevisiae SEC4 signal transduction 12 x DUSP3 dual specificity phosphatase 3 signal transduction 11 x NOLA2 nucleolar protein family A, member 2 protein syntesis 11 x RAB11A RAB11A, member RAS oncogene family signal transduction 10 x The classifiers at the best Lauren filtering level Genetic reducer 3
Lauren Lauren 1R Classifier Dynamic reducer Lymph node metastasis Lymph node metastasis Genetic reducer 1R Classifier 4
Comparison of the learning methods Conclusions using the best discretization method • Genes function in teams: Identification of individual, significantly differentially expressed genes only is insufficient for classification • Rough set learning identifies different groups of genes for different objects • RS learning outperforms both linear and quadratic linear discriminant analysis • Literature validation could not be completed; present knowledge of cancer is scarce and fragmented • RS supervised learning provides valuable hypotheses about molecular functions of genes • Combination of rough sets with feature selection methods may be well suited for this task • But: only 2,504 genes out of at least 30K genes were Lauren used 5
Recommend
More recommend