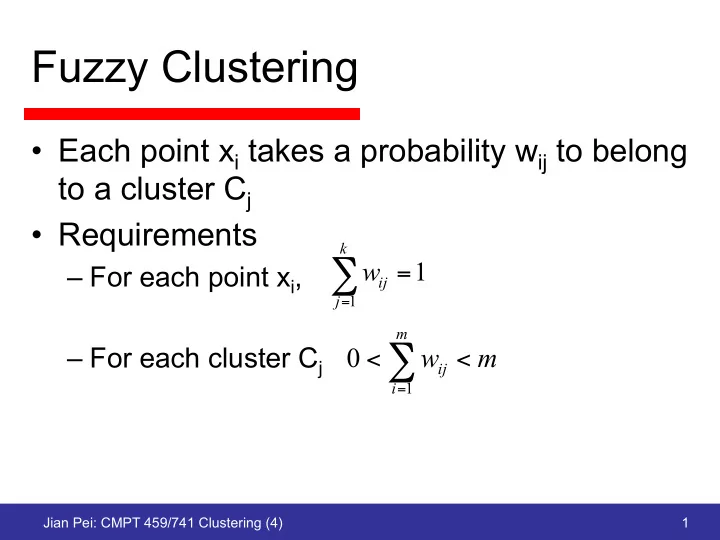

Fuzzy Clustering • Each point x i takes a probability w ij to belong to a cluster C j • Requirements k w 1 ∑ – For each point x i , = ij j 1 = m – For each cluster C j 0 w m < ∑ ij < i = 1 Jian Pei: CMPT 459/741 Clustering (4) 1
Fuzzy C-Means (FCM) Select an initial fuzzy pseudo-partition, i.e., assign values to all the w ij Repeat Compute the centroid of each cluster using the fuzzy pseudo-partition Recompute the fuzzy pseudo-partition, i.e., the w ij Until the centroids do not change (or the change is below some threshold) Jian Pei: CMPT 459/741 Clustering (4) 2
Critical Details • Optimization on sum of the squared error (SSE): k m p 2 SSE ( C , … , C ) w dist ( x , c ) ∑∑ = 1 k ij i j j 1 i 1 = = m m p p c w x / w • Computing centroids: ∑ ∑ = j ij i ij i 1 i 1 = = • Updating the fuzzy pseudo-partition 1 1 k 2 p 1 2 p 1 w ( 1 / dist ( x , c ) ) ( 1 / dist ( x , c ) ) − − ∑ = ij i j i q q 1 = k – When p=2 2 2 w 1 / dist ( x , c ) 1 / dist ( x , c ) ∑ = ij i j i q q 1 = Jian Pei: CMPT 459/741 Clustering (4) 3
Choice of P • When p à 1, FCM behaves like traditional k-means • When p is larger, the cluster centroids approach the global centroid of all data points • The partition becomes fuzzier as p increases Jian Pei: CMPT 459/741 Clustering (4) 4
Effectiveness Jian Pei: CMPT 459/741 Clustering (4) 5
Mixture Models • A cluster can be modeled as a probability distribution – Practically, assume a distribution can be approximated well using multivariate normal distribution • Multiple clusters is a mixture of different probability distributions • A data set is a set of observations from a mixture of models Jian Pei: CMPT 459/741 Clustering (4) 6
Object Probability • Suppose there are k clusters and a set X of m objects – Let the j-th cluster have parameter θ j = ( µ j , σ j ) – The probability that a point is in the j-th cluster is w j , w 1 + … + w k = 1 • The probability of an object x is k prob ( x | ) w p ( x | ) ∑ Θ = θ j j j j 1 = m m k prob ( X | ) prob ( x | ) w p ( x | ) ∏ ∏∑ Θ = Θ = θ i j j i j i 1 i 1 j 1 = = = Jian Pei: CMPT 459/741 Clustering (4) 7
Example 2 ( x ) − µ 1 − 2 prob ( x | ) e 2 σ Θ = i 2 π σ ( 4 , 2 ) ( 4 , 2 ) θ = − θ = 1 2 2 2 ( x 4 ) ( x 4 ) + − 1 1 − − prob ( x | ) e e 8 8 Θ = + 2 2 2 2 π π Jian Pei: CMPT 459/741 Clustering (4) 8
Maximal Likelihood Estimation • Maximum likelihood principle: if we know a set of objects are from one distribution, but do not know the parameter, we can choose the parameter maximizing the probability 2 ( x ) • Maximize − µ m 1 − 2 prob ( x | ) e 2 ∏ Θ = σ i 2 π σ j 1 = – Equivalently, maximize 2 m ( x ) − µ log prob ( X | ) i 0 . 5 m log 2 m log ∑ Θ = − − π − σ 2 2 σ i 1 = Jian Pei: CMPT 459/741 Clustering (4) 9
EM Algorithm • Expectation Maximization algorithm Select an initial set of model parameters Repeat Expectation Step: for each object, calculate the probability that it belongs to each distribution θ i , i.e., prob(x i | θ i ) Maximization Step: given the probabilities from the expectation step, find the new estimates of the parameters that maximize the expected likelihood Until the parameters are stable Jian Pei: CMPT 459/741 Clustering (4) 10
Advantages and Disadvantages • Mixture models are more general than k- means and fuzzy c-means • Clusters can be characterized by a small number of parameters • The results may satisfy the statistical assumptions of the generative models • Computationally expensive • Need large data sets • Hard to estimate the number of clusters Jian Pei: CMPT 459/741 Clustering (4) 11
Grid-based Clustering Methods • Ideas – Using multi-resolution grid data structures – Using dense grid cells to form clusters • Several interesting methods – CLIQUE – STING – WaveCluster Jian Pei: CMPT 459/741 Clustering (4) 12
CLIQUE • Clustering In QUEst • Automatically identify subspaces of a high dimensional data space • Both density-based and grid-based Jian Pei: CMPT 459/741 Clustering (4) 13
CLIQUE: the Ideas • Partition each dimension into the same number of equal length intervals – Partition an m-dimensional data space into non- overlapping rectangular units • A unit is dense if the number of data points in the unit exceeds a threshold • A cluster is a maximal set of connected dense units within a subspace Jian Pei: CMPT 459/741 Clustering (4) 14
CLIQUE: the Method • Partition the data space and find the number of points in each cell of the partition – Apriori: a k-d cell cannot be dense if one of its (k-1)-d projection is not dense • Identify clusters: – Determine dense units in all subspaces of interests and connected dense units in all subspaces of interests • Generate minimal description for the clusters – Determine the minimal cover for each cluster Jian Pei: CMPT 459/741 Clustering (4) 15
CLIQUE: An Example 6 7 (10,000) Salary 5 Vac atio n 4 3 30 50 1 2 age age 0 Vacation (week) 20 30 40 50 60 6 7 5 4 3 1 2 age 0 20 30 40 50 60 Jian Pei: CMPT 459/741 Clustering (4) 16
CLIQUE: Pros and Cons • Automatically find subspaces of the highest dimensionality with high density clusters • Insensitive to the order of input – Not presume any canonical data distribution • Scale linearly with the size of input • Scale well with the number of dimensions • The clustering result may be degraded at the expense of simplicity of the method Jian Pei: CMPT 459/741 Clustering (4) 17
Bad Cases for CLIQUE Parts of a cluster may be missed A cluster from CLIQUE may contain noise Jian Pei: CMPT 459/741 Clustering (4) 18
Dimensionality Reduction • Clustering a high dimensional data set is challenging – Distance between two points could be dominated by noise • Dimensionality reduction: choosing the informative dimensions for clustering analysis – Feature selection: choosing a subset of existing dimensions – Feature construction: construct a new (small) set of informative attributes Jian Pei: CMPT 459/741 Clustering (4) 19
Variance and Covariance • Given a set of 1-d points, how different are those points? n 2 ( X X ) = ∑ − i – Standard deviation: s i 1 = n n 1 − 2 ( X X ) = ∑ − – Variance: i 2 s i 1 = n 1 − • Given a set of 2-d points, are the two dimensions correlated? n – Covariance: ( X X )( Y Y ) = ∑ − − i i i 1 cov( X , Y ) = n 1 − Jian Pei: CMPT 459/741 Clustering (4) 20
Principal Components Art work and example from http://csnet.otago.ac.nz/cosc453/student_tutorials/principal_components.pdf Jian Pei: CMPT 459/741 Clustering (4) 21
Step 1: Mean Subtraction • Subtract the mean from each dimension for each data point • Intuition: centralizing the data set Jian Pei: CMPT 459/741 Clustering (4) 22
Step 2: Covariance Matrix � cov( D , D ) cov( D , D ) cov( D , D ) ⎛ ⎞ 1 1 1 2 1 n ⎜ ⎟ cov( D , D ) cov( D , D ) � cov( D , D ) ⎜ ⎟ 2 1 2 2 2 n C = ⎜ ⎟ � � � � ⎜ ⎟ ⎜ ⎟ cov( D , D ) cov( D , D ) � cov( D , D ) ⎝ ⎠ n 1 n 2 n n Jian Pei: CMPT 459/741 Clustering (4) 23
Step 3: Eigenvectors and Eigenvalues • Compute the eigenvectors and the eigenvalues of the covariance matrix – Intuition: find those direction invariant vectors as candidates of new attributes – Eigenvalues indicate how much the direction invariant vectors are scaled – the larger the better for manifest the data variance Jian Pei: CMPT 459/741 Clustering (4) 24
Step 4: Forming New Features • Choose the principal components and forme new features – Typically, choose the top-k components Jian Pei: CMPT 459/741 Clustering (4) 25
New Features NewData = RowFeatureVector x RowDataAdjust The first principal component is used Jian Pei: CMPT 459/741 Clustering (4) 26
Clustering in Derived Space Y - 0.707x + 0.707y O X Jian Pei: CMPT 459/741 Clustering (4) 27
Spectral Clustering Data Affinity matrix Computing the leading Clustering in the Projecting back to k eigenvectors of A new space cluster the original data [ ] W ij Av = \lamda v A = f(W) Jian Pei: CMPT 459/741 Clustering (4) 28
Affinity Matrix • Using a distance measure dist ( oi,oj ) W ij = e − σ w where σ is a scaling parameter controling how fast the affinity W ij decreases as the distance increases • In the Ng-Jordan-Weiss algorithm, W ii is set to 0 Jian Pei: CMPT 459/741 Clustering (4) 29
Clustering • In the Ng-Jordan-Weiss algorithm, we define a diagonal matrix such that n X D ii = W ij j =1 • Then, A = D − 1 2 WD − 1 2 • Use the k leading eigenvectors to form a new space • Map the original data to the new space and conduct clustering Jian Pei: CMPT 459/741 Clustering (4) 30
Recommend
More recommend