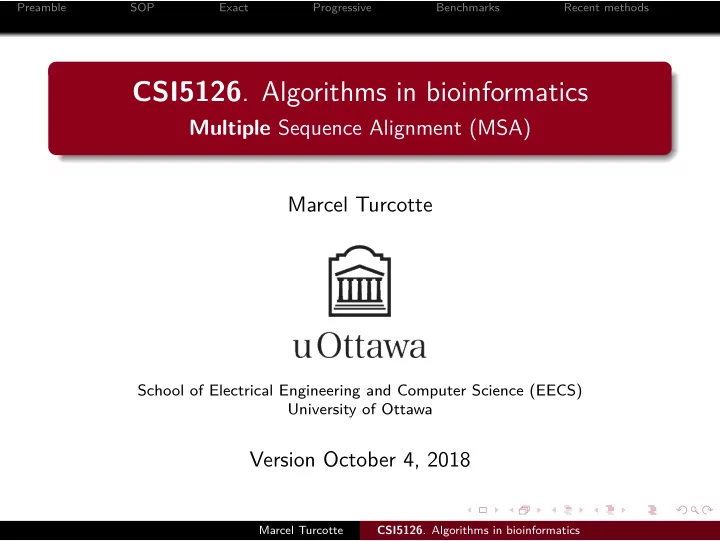

. d2rmbi_ KGSCFHRIIPGFMCQGGDFTRHNGTGGKSIYGEKFEDENFILKHTGPGILSMANAGPNTN d1rmha_ KGSCFHRIIPGFMCQGGDFTRHNGTGGKSIYGEKFEDENFILKHTGPGILSMANAGPNTN d1ak4b_ KGSCFHRIIPGFMCQGGDFTRHNGTGGKSIYGEKFEDENFILKHTGPGILSMANAGPNTN d2rmbc_ KGSCFHRIIPGFMCQGGDFTRHNGTGGKSIYGEKFEDENFILKHTGPGILSMANAGPNTN Now what ? KGSCFHRIIPGFXCQGGDFTRHNGTGGKSIYGEKFEDENFILKHTGPGILSXANAGPNTN Motivation Recent methods Benchmarks Progressive Exact SOP Preamble Recent methods d1awtf_ d1cyna_ Progressive * Marcel Turcotte ... . : . . . .. : KNSKFHRVIKDFMIQGGDFTRGDGTGGKSIYGERFPDENFKLKHYGPGWVSMANAGKDTN ------AIDKPFLLPIEDVFSISGRG--TVVTGRVERGIIKVGEEVEIVGIKETQKSTCT d1efca1 NNTTFHRVIPGFMIQGGGFTEQMQQ--KKPNPPIKNEADNGLRNTRGTIAMARTADKDSA d1clh__ KGSTFHRVIKNFMIQGGDFTKGDGTGGESIYGGMFDDEEFVMKHDEPFVVSMANKGPNTN d1c5fg_ KGSIFHRVIKDFMIQGGDFTARDGTGGMSIYGETFPDENFKLKHYGIGWVSMANAGPDTN d2rmce_ Benchmarks Exact . . . . . . . . . . . . . . . . . . . . . SOP . Preamble . . . . . . . . . . . . . . . . . CSI5126 . Algorithms in bioinformatics
. d2rmbi_ KGSCFHRIIPGFMCQGGDFTRHNGTGGKSIYGEKFEDENFILKHTGPGILSMANAGPNTN d1rmha_ KGSCFHRIIPGFMCQGGDFTRHNGTGGKSIYGEKFEDENFILKHTGPGILSMANAGPNTN d1ak4b_ KGSCFHRIIPGFMCQGGDFTRHNGTGGKSIYGEKFEDENFILKHTGPGILSMANAGPNTN d2rmbc_ KGSCFHRIIPGFMCQGGDFTRHNGTGGKSIYGEKFEDENFILKHTGPGILSMANAGPNTN Now what ? KGSCFHRIIPGFXCQGGDFTRHNGTGGKSIYGEKFEDENFILKHTGPGILSXANAGPNTN Motivation Recent methods Benchmarks Progressive Exact SOP Preamble Recent methods d1awtf_ d1cyna_ Progressive * Marcel Turcotte ... . : . . . .. : KNSKFHRVIKDFMIQGGDFTRGDGTGGKSIYGERFPDENFKLKHYGPGWVSMANAGKDTN ------AIDKPFLLPIEDVFSISGRG--TVVTGRVERGIIKVGEEVEIVGIKETQKSTCT d1efca1 NNTTFHRVIPGFMIQGGGFTEQMQQ--KKPNPPIKNEADNGLRNTRGTIAMARTADKDSA d1clh__ KGSTFHRVIKNFMIQGGDFTKGDGTGGESIYGGMFDDEEFVMKHDEPFVVSMANKGPNTN d1c5fg_ KGSIFHRVIKDFMIQGGDFTARDGTGGMSIYGETFPDENFKLKHYGIGWVSMANAGPDTN d2rmce_ Benchmarks Exact . . . . . . . . . . . . . . . . . . . . . SOP . Preamble . . . . . . . . . . . . . . . . . CSI5126 . Algorithms in bioinformatics
. Preamble . . . . . . Preamble SOP Exact Progressive Benchmarks Recent methods SOP . Exact Progressive Benchmarks Recent methods Computer Science ’s Point of View: Generalization An MSA ( multiple sequence alignment ) is a generalization of the pairwise sequence alignment. gaps are inserted so that 1) all the sequences have the same length and 2) the distance for the alignment is minimized (this can also be seen as to maximize the similarity). Global or local multiple alignment. Marcel Turcotte . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . CSI5126 . Algorithms in bioinformatics Defjnition. Given k > 2 strings S = { S 1 , S 2 , . . . , S k } ,
. Exact . . . . . . . . . Preamble SOP Progressive . Benchmarks Recent methods Preamble SOP Exact Progressive Benchmarks Recent methods Life Science ’s Point of View Conserved patterns , e.g. conserved cysteins forming disulphide bonds . Marcel Turcotte . . . . . . . . . . . . . . . . . . . . . . . . . . . . . CSI5126 . Algorithms in bioinformatics
. Exact . . . . . . . . . Preamble SOP Progressive . Benchmarks Recent methods Preamble SOP Exact Progressive Benchmarks Recent methods Life Science ’s Point of View Conserved Pro and Gl y opposed to an insersion suggest the Marcel Turcotte . . . . . . . . . . . . . . . . . . . . . . . . . . . . . CSI5126 . Algorithms in bioinformatics presence of a loop .
. Exact . . . . . . . . . Preamble SOP Progressive . Benchmarks Recent methods Preamble SOP Exact Progressive Benchmarks Recent methods Life Science ’s Point of View Similarity . Marcel Turcotte . . . . . . . . . . . . . . . . . . . . . . . . . . . . . CSI5126 . Algorithms in bioinformatics
. Exact . . . . . . . . . Preamble SOP Progressive . Benchmarks Recent methods Preamble SOP Exact Progressive Benchmarks Recent methods Life Science ’s Point of View Chemical properties . Marcel Turcotte . . . . . . . . . . . . . . . . . . . . . . . . . . . . . CSI5126 . Algorithms in bioinformatics
. Exact . . . . . . . . . Preamble SOP Progressive . Benchmarks Recent methods Preamble SOP Exact Progressive Benchmarks Recent methods Life Science ’s Point of View Patterns of conservation/substitution can indicate a preference for solvent exposure . Marcel Turcotte . . . . . . . . . . . . . . . . . . . . . . . . . . . . . CSI5126 . Algorithms in bioinformatics
. Exact . . . . . . . . . Preamble SOP Progressive . Benchmarks Recent methods Preamble SOP Exact Progressive Benchmarks Recent methods Life Science ’s Point of View Secondary structure elements? Marcel Turcotte . . . . . . . . . . . . . . . . . . . . . . . . . . . . . CSI5126 . Algorithms in bioinformatics
. Exact . . . . . . . . . Preamble SOP Progressive . Benchmarks Recent methods Preamble SOP Exact Progressive Benchmarks Recent methods Life Science ’s Point of View Gaps are good indicators of loop regions. Marcel Turcotte . . . . . . . . . . . . . . . . . . . . . . . . . . . . . CSI5126 . Algorithms in bioinformatics
. Exact . . . . . . . . . Preamble SOP Progressive . Benchmarks Recent methods Preamble SOP Exact Progressive Benchmarks Recent methods Life Science ’s Point of View History (phylogeny) Marcel Turcotte . . . . . . . . . . . . . . . . . . . . . . . . . . . . . CSI5126 . Algorithms in bioinformatics
. SOP . . . . . . . . . . Preamble Exact . Progressive Benchmarks Recent methods Preamble SOP Exact Progressive Benchmarks Recent methods Life Science ’s Point of View Marcel Turcotte . . . . . . . . . . . . . . . . . . . . . . . . . . . . CSI5126 . Algorithms in bioinformatics
. Recent methods . . . . . . . Preamble SOP Exact Progressive Benchmarks Preamble . SOP Exact Progressive Benchmarks Recent methods Life Science ’s Point of View “ Multiple alignments are among the most useful objects in bioinformatics ” [Wallace 2005] Phylogenetic trees inference Identifying functional residues Structure prediction etc. Marcel Turcotte . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . CSI5126 . Algorithms in bioinformatics
. Benchmarks . . . . . . . . Preamble SOP Exact Progressive Recent methods . Preamble SOP Exact Progressive Benchmarks Recent methods Pairwise vs Multiple Sequence Alignment Pairwise : the question is “ are the two sequences related? ” Multiple : the sequences are assumed to be related from the start. Marcel Turcotte . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . CSI5126 . Algorithms in bioinformatics
. Benchmarks . . . . . . . . Preamble SOP Exact Progressive Recent methods . Preamble SOP Exact Progressive Benchmarks Recent methods Pairwise vs Multiple Sequence Alignment Pairwise : the question is “ are the two sequences related? ” Multiple : the sequences are assumed to be related from the start. Marcel Turcotte . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . CSI5126 . Algorithms in bioinformatics
. Exact . . . . . Preamble SOP Exact Progressive Benchmarks Recent methods Preamble SOP Progressive . Benchmarks Recent methods Multiple Sequence Alignment Rectangular table such that: Rows are (related, homologous) sequences Residues in a given column (site): 1. Evolved from a position in some ancestral sequence (homologous) 2. Can be superimposed in three-dimension in a structural alignment 3. Have the same functional role All three criteria might not be simultaneously met, especially for sequences that are not closely related. Marcel Turcotte . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . CSI5126 . Algorithms in bioinformatics
. Exact . . . . . Preamble SOP Exact Progressive Benchmarks Recent methods Preamble SOP Progressive . Benchmarks Recent methods Multiple Sequence Alignment Rectangular table such that: Rows are (related, homologous) sequences Residues in a given column (site): 1. Evolved from a position in some ancestral sequence (homologous) 2. Can be superimposed in three-dimension in a structural alignment 3. Have the same functional role All three criteria might not be simultaneously met, especially for sequences that are not closely related. Marcel Turcotte . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . CSI5126 . Algorithms in bioinformatics
. Exact . . . . . Preamble SOP Exact Progressive Benchmarks Recent methods Preamble SOP Progressive . Benchmarks Recent methods Multiple Sequence Alignment Rectangular table such that: Rows are (related, homologous) sequences Residues in a given column (site): 1. Evolved from a position in some ancestral sequence (homologous) 2. Can be superimposed in three-dimension in a structural alignment 3. Have the same functional role All three criteria might not be simultaneously met, especially for sequences that are not closely related. Marcel Turcotte . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . CSI5126 . Algorithms in bioinformatics
. Exact . . . . . Preamble SOP Exact Progressive Benchmarks Recent methods Preamble SOP Progressive . Benchmarks Recent methods Multiple Sequence Alignment Rectangular table such that: Rows are (related, homologous) sequences Residues in a given column (site): 1. Evolved from a position in some ancestral sequence (homologous) 2. Can be superimposed in three-dimension in a structural alignment 3. Have the same functional role All three criteria might not be simultaneously met, especially for sequences that are not closely related. Marcel Turcotte . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . CSI5126 . Algorithms in bioinformatics
. Exact . . . . . Preamble SOP Exact Progressive Benchmarks Recent methods Preamble SOP Progressive . Benchmarks Recent methods Multiple Sequence Alignment Rectangular table such that: Rows are (related, homologous) sequences Residues in a given column (site): 1. Evolved from a position in some ancestral sequence (homologous) 2. Can be superimposed in three-dimension in a structural alignment 3. Have the same functional role All three criteria might not be simultaneously met, especially for sequences that are not closely related. Marcel Turcotte . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . CSI5126 . Algorithms in bioinformatics
. Benchmarks 1 BLOSUM62. k n Objective function : sum-of-pairs Recent methods Progressive n Exact SOP Preamble Recent methods . Progressive Exact c 1 Preamble LGB2LUPLU Marcel Turcotte sum-of-pairs (SP) score. Problem: Compute the (global) alignment that maximizes the ...IAGADNGAGV... GLB1GLYDI k ...FNA--NIPKH... ...VKG------D... HBA_HUMAN j-> GLB2HCHITP ...VEA--DVAGH... MYG_PHYCA i-> ...V----NVDEV... HBB_HUMAN ...VGA--HAGEY... SOP Benchmarks . . . . . . . . . . . . . . . . . . . . . . . . CSI5126 . Algorithms in bioinformatics . . . . . . . . . . . . . . Given a multiple alignment M of k sequences and n columns. k − 1 ∑ ∑ ∑ sp ( M ) = s ( m ci , m cj ) c = 1 i = 1 j = i + 1 where s ( a , b ) is a substitution matrix such as PAM250 or
. Benchmarks . . . . . . . . Preamble SOP Exact Progressive Recent methods . Preamble SOP Exact Progressive Benchmarks Recent methods Remarks Unlike pairwise aligment, the sum-of-pairs score used by the MSA methods has no theoretical fundation , no interpretation in terms of an underlying evolutionary model. Marcel Turcotte . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . CSI5126 . Algorithms in bioinformatics
. SOP . . . . . . . . . . Preamble Exact . Progressive Benchmarks Recent methods Preamble SOP Exact Progressive Benchmarks Recent methods Sum of pairs Marcel Turcotte . . . . . . . . . . . . . . . . . . . . . . CSI5126 . Algorithms in bioinformatics . . . . . . C C C A C C C T A C A A T C T T
. Progressive . . . . Preamble SOP Exact Progressive Benchmarks Recent methods Preamble SOP Exact Benchmarks . Recent methods Remarks I. M. Wallace, G. Blackshields, and D. G. Higgins. Multiple sequence alignments. Curr Opin Struct Biol , 15(3):261–6, Jun 2005. “Assembling a suitable MSA is not, however, a trivial task, and none of the existing methods have yet managed to deliver biologically perfect MSAs. ” “ Manually refjned alignments continue to be superior to purely automated methods;” “The wealth of available methods and their increasingly similar accuracies makes it harder than ever to objectively choose one over the others.” Marcel Turcotte . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . CSI5126 . Algorithms in bioinformatics
. Progressive . . . . . . . . Preamble SOP Exact Benchmarks . Recent methods Preamble SOP Exact Progressive Benchmarks Recent methods 2 , 3 , k , go ! Optimal alignment of 2 sequences Optimal alignment of 3 sequences Optimal alignment of k sequences Marcel Turcotte . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . CSI5126 . Algorithms in bioinformatics
. SOP . . . . . . . . . . Preamble Exact . Progressive Benchmarks Recent methods Preamble SOP Exact Progressive Benchmarks Recent methods Optimal alignment of 2 Sequences Marcel Turcotte . . . . . . . . . . . . . . . . . . . . . . . . . . . . CSI5126 . Algorithms in bioinformatics V V
. Exact . . . . . . . . . Preamble SOP Progressive . Benchmarks Recent methods Preamble SOP Exact Progressive Benchmarks Recent methods Optimal alignment of 2 Sequences V V Marcel Turcotte . . . . . . . . . . . . . . . . . . . . . . . . . . . . . CSI5126 . Algorithms in bioinformatics V V
. Exact . . . . . . . . . Preamble SOP Progressive . Benchmarks Recent methods Preamble SOP Exact Progressive Benchmarks Recent methods Optimal alignment of 2 Sequences V - Marcel Turcotte . . . . . . . . . . . . . . . . . . . . . . . . . . . . . CSI5126 . Algorithms in bioinformatics V W
. Exact . . . . . . . . . Preamble SOP Progressive . Benchmarks Recent methods Preamble SOP Exact Progressive Benchmarks Recent methods Optimal alignment of 2 Sequences - W Marcel Turcotte . . . . . . . . . . . . . . . . . . . . . . . . . . . . . CSI5126 . Algorithms in bioinformatics V W
. SOP . . . . . . . . . . Preamble Exact . Progressive Benchmarks Recent methods Preamble SOP Exact Progressive Benchmarks Recent methods Optimal alignment of 3 Sequences Marcel Turcotte . . . . . . . . . . . . . . . . . . . . . . . . . . . . CSI5126 . Algorithms in bioinformatics V V V
. Progressive . . . . . . . . Preamble SOP Exact Benchmarks . Recent methods Preamble SOP Exact Progressive Benchmarks Recent methods Optimal alignment of 3 Sequences V V V Marcel Turcotte . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . CSI5126 . Algorithms in bioinformatics V V V
. Progressive . . . . . . . . Preamble SOP Exact Benchmarks . Recent methods Preamble SOP Exact Progressive Benchmarks Recent methods Optimal alignment of 3 Sequences V V - Marcel Turcotte . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . CSI5126 . Algorithms in bioinformatics W V V
. Progressive . . . . . . . . Preamble SOP Exact Benchmarks . Recent methods Preamble SOP Exact Progressive Benchmarks Recent methods Optimal alignment of 3 Sequences - V V Marcel Turcotte . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . CSI5126 . Algorithms in bioinformatics V W V
. Progressive . . . . . . . . Preamble SOP Exact Benchmarks . Recent methods Preamble SOP Exact Progressive Benchmarks Recent methods Optimal alignment of 3 Sequences V - V Marcel Turcotte . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . CSI5126 . Algorithms in bioinformatics V V W
. Progressive . . . . . . . . Preamble SOP Exact Benchmarks . Recent methods Preamble SOP Exact Progressive Benchmarks Recent methods Optimal alignment of 3 Sequences V - - Marcel Turcotte . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . CSI5126 . Algorithms in bioinformatics L V W
. Progressive . . . . . . . . Preamble SOP Exact Benchmarks . Recent methods Preamble SOP Exact Progressive Benchmarks Recent methods Optimal alignment of 3 Sequences - W - Marcel Turcotte . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . CSI5126 . Algorithms in bioinformatics L V W
. Progressive . . . . . . . . Preamble SOP Exact Benchmarks . Recent methods Preamble SOP Exact Progressive Benchmarks Recent methods Optimal alignment of 3 Sequences - - L Marcel Turcotte . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . CSI5126 . Algorithms in bioinformatics L V W
. SOP . . . . . . . . . . Preamble Exact . Progressive Benchmarks Recent methods Preamble SOP Exact Progressive Benchmarks Recent methods Optimal alignment of 3 Sequences Marcel Turcotte . . . . . . . . . . . . . . . . . . . . . . . . . . . . CSI5126 . Algorithms in bioinformatics
S N G N S G N A . . . . . .. . . . . . . .. . . . . . . .. . . . . . . .. . . .. . . . . .
G G - S N G N S G N A . . . . . .. . . . . . . .. . . . . . . .. . . . . . . .. . . .. . . . . .
GN GN -N S N G N S G N A . . . . . .. . . . . . . .. . . . . . . .. . . . . . . .. . . .. . . . . .
GN- GNA -N- S N G N S G N A . . . . . .. . . . . . . .. . . . . . . .. . . . . . . .. . . .. . . . . .
GN-S GNA- -N-S S N G N S G N A . . . . . .. . . . . . . .. . . . . . . .. . . . . . . .. . . .. . . . . .
. Exact . . . . . . . . . Preamble SOP Progressive . Benchmarks . Preamble SOP Exact Progressive Benchmarks Recent methods Exact alignment of 3 Sequences Given : x , y and z three strings. given by: Marcel Turcotte . Recent methods . . . . . . . . . . . . . . CSI5126 . Algorithms in bioinformatics . . . . . . . . . . . . . V ( i , j , k ) is the optimal SP score to align x [ 1 .. i ] , y [ 1 .. j ] and z [ 1 .. k ] is V ( i − 1 , j − 1 , k − 1 ) + s ( x i , y j , z k ) , V ( i − 1 , j − 1 , k ) − s ( x i , y j , − ) , V ( i , j − 1 , k − 1 ) − s ( − , y j , z k ) , V ( i , j , k ) = max V ( i − 1 , j , k − 1 ) − s ( x i , − , z k ) , V ( i − 1 , j , k ) − s ( x i , − , − ) , V ( i , j − 1 , k ) − s ( − , y j , − ) , V ( i , j , k − 1 ) − s ( − , − , z k ) . ⇒ For non-boundary cells only.
. Exact . . . . . . . . . Preamble SOP Progressive . Benchmarks Recent methods Preamble SOP Exact Progressive Benchmarks Recent methods Exact alignment of 3 Sequences At the boundaries: Marcel Turcotte . . . . . . . . . . . . . . . . . . . . . . . CSI5126 . Algorithms in bioinformatics . . . . . . V ( 0 , 0 , 0 ) = 0 , V ( i , j , 0 ) = V ( x i , y j ) − ( i + j ) × d , V ( i , 0 , k ) = V ( x i , z k ) − ( i + k ) × d , V ( 0 , j , k ) = V ( y j , z k ) − ( j + k ) × d .
. SOP . . . . . . . . . . Preamble Exact . Progressive Benchmarks Recent methods . SOP Exact Progressive Benchmarks Recent methods Exact alignment of k Sequences Marcel Turcotte . Preamble . . . . . . . . . . . . . CSI5126 . Algorithms in bioinformatics . . . . . . . . . . . . . Given : x 1 , x 2 and x k , k sequences. The optimum SP alignment for k sequences, V ( i 1 , i 2 , . . . , i k ) , to align x 1 [ 1 .. i 1 ] , x 2 [ 1 .. i 2 ] , . . . , x k [ 1 .. i k ] V ( i 1 − 1 , i 2 − 1 , . . . , i k − 1 ) + s ( i 1 , i 2 , . . . , i k ) , V ( i 1 , i 2 − 1 , . . . , i k − 1 ) + s ( − , i 2 , . . . , i k ) , V ( i 1 − 1 , i 2 , . . . , i k − 1 ) + s ( i 1 , − , . . . , i k ) , . . . V ( i 1 , i 2 , . . . , i k ) = max V ( i 1 − 1 , i 2 − 1 , . . . , i k ) + s ( i 1 , i 2 , . . . , − ) , . . . V ( i 1 , i 2 , . . . , i k − 1 ) + s ( − , − , . . . , i k ) , . . . ⇒ All the subsets (2 k ) except the empty one, which corresponds to − , − , . . . , − , hence, 2 k − 1 cases.
O n k (memory cells) Time complexity is O 2 k n k Say k . Benchmarks Space complexity is Given: k sequences of approximately the same length, n Recall that peta- (P) = 10 15 , tera- (T) 10 12 , giga- (G) 10 9 Remarks Recent methods Progressive 100 and k Exact SOP Preamble Recent methods Benchmarks Progressive Exact For n 10 10 5, n k 5, n Marcel Turcotte 16 million years! 100 takes 10, n For k 26 hours 100 takes For k Preamble 100 takes 0.2 millisecond n 2 (pairwise) and 10 20 10, n k 100 and k For n SOP . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . CSI5126 . Algorithms in bioinformatics
O n k (memory cells) Time complexity is O 2 k n k Say k . Benchmarks Space complexity is Given: k sequences of approximately the same length, n Recall that peta- (P) = 10 15 , tera- (T) 10 12 , giga- (G) 10 9 Remarks Recent methods Progressive 100 and k Exact SOP Preamble Recent methods Benchmarks Progressive Exact For n 10 10 5, n k 5, n Marcel Turcotte 16 million years! 100 takes 10, n For k 26 hours 100 takes For k Preamble 100 takes 0.2 millisecond n 2 (pairwise) and 10 20 10, n k 100 and k For n SOP . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . CSI5126 . Algorithms in bioinformatics
Time complexity is O 2 k n k Say k O n k (memory cells) . Benchmarks Space complexity is Given: k sequences of approximately the same length, n Recall that peta- (P) = 10 15 , tera- (T) 10 12 , giga- (G) 10 9 Remarks Recent methods Progressive 100 and k Exact SOP Preamble Recent methods Benchmarks Progressive Exact For n 10 10 5, n k 5, n Marcel Turcotte 16 million years! 100 takes 10, n For k 26 hours 100 takes For k Preamble 100 takes 0.2 millisecond n 2 (pairwise) and 10 20 10, n k 100 and k For n SOP . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . CSI5126 . Algorithms in bioinformatics
Time complexity is O 2 k n k Say k O n k (memory cells) . Benchmarks Space complexity is Given: k sequences of approximately the same length, n Recall that peta- (P) = 10 15 , tera- (T) 10 12 , giga- (G) 10 9 Remarks Recent methods Progressive 100 and k Exact SOP Preamble Recent methods Benchmarks Progressive Exact For n 10 10 5, n k 5, n Marcel Turcotte 16 million years! 100 takes 10, n For k 26 hours 100 takes For k Preamble 100 takes 0.2 millisecond n 2 (pairwise) and 10 20 10, n k 100 and k For n SOP . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . CSI5126 . Algorithms in bioinformatics
O 2 k n k Say k . Exact Recall that peta- (P) = 10 15 , tera- (T) 10 12 , giga- (G) 10 9 Remarks Recent methods Benchmarks Progressive SOP For n Preamble Recent methods Benchmarks Progressive Exact SOP Preamble Given: k sequences of approximately the same length, n 100 and k . 100 takes Marcel Turcotte 16 million years! 100 takes 10, n For k 26 hours 5, n 10, n k For k 100 takes 0.2 millisecond n 2 (pairwise) and Time complexity is 10 20 . . . . . . . . . . . . . . . . . . . . . CSI5126 . Algorithms in bioinformatics . . . . . . . . . . . . . . . . . Space complexity is O ( n k ) (memory cells) For n = 100 and k = 5, n k = 10 10
O 2 k n k Say k . Recent methods Benchmarks Progressive Exact SOP Preamble Benchmarks Remarks Progressive Exact SOP Preamble . . Recent methods Recall that peta- (P) = 10 15 , tera- (T) 10 12 , giga- (G) 10 9 . Given: k sequences of approximately the same length, n Time complexity is 2 (pairwise) and n 100 takes 0.2 millisecond For k 5, n 100 takes 26 hours For k 10, n 100 takes 16 million years! Marcel Turcotte . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . CSI5126 . Algorithms in bioinformatics Space complexity is O ( n k ) (memory cells) For n = 100 and k = 5, n k = 10 10 For n = 100 and k = 10, n k = 10 20
O 2 k n k Say k . Recent methods Benchmarks Progressive Exact SOP Preamble Benchmarks Remarks Progressive Exact SOP Preamble . . Recent methods Recall that peta- (P) = 10 15 , tera- (T) 10 12 , giga- (G) 10 9 . Given: k sequences of approximately the same length, n Time complexity is 2 (pairwise) and n 100 takes 0.2 millisecond For k 5, n 100 takes 26 hours For k 10, n 100 takes 16 million years! Marcel Turcotte . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . CSI5126 . Algorithms in bioinformatics Space complexity is O ( n k ) (memory cells) For n = 100 and k = 5, n k = 10 10 For n = 100 and k = 10, n k = 10 20
O 2 k n k Say k . Recent methods Benchmarks Progressive Exact SOP Preamble Benchmarks Remarks Progressive Exact SOP Preamble . . Recent methods Recall that peta- (P) = 10 15 , tera- (T) 10 12 , giga- (G) 10 9 . Given: k sequences of approximately the same length, n Time complexity is 2 (pairwise) and n 100 takes 0.2 millisecond For k 5, n 100 takes 26 hours For k 10, n 100 takes 16 million years! Marcel Turcotte . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . CSI5126 . Algorithms in bioinformatics Space complexity is O ( n k ) (memory cells) For n = 100 and k = 5, n k = 10 10 For n = 100 and k = 10, n k = 10 20
. Recent methods . . . . . . . Preamble SOP Exact Progressive Benchmarks Preamble . SOP Exact Progressive Benchmarks Recent methods Remarks Recall that peta- (P) = 10 15 , tera- (T) 10 12 , giga- (G) 10 9 Given: k sequences of approximately the same length, n For k 10, n 100 takes 16 million years! Marcel Turcotte . . . . . . . . . . . . . . . . . . . . . . . . CSI5126 . Algorithms in bioinformatics . . . . . . . Space complexity is O ( n k ) (memory cells) For n = 100 and k = 5, n k = 10 10 For n = 100 and k = 10, n k = 10 20 Time complexity is O ( 2 k n k ) Say k = 2 (pairwise) and n = 100 takes 0.2 millisecond For k = 5, n = 100 takes ∼ 26 hours
. Recent methods . . . . . . . Preamble SOP Exact Progressive Benchmarks Preamble . SOP Exact Progressive Benchmarks Recent methods Remarks Recall that peta- (P) = 10 15 , tera- (T) 10 12 , giga- (G) 10 9 Given: k sequences of approximately the same length, n For k 10, n 100 takes 16 million years! Marcel Turcotte . . . . . . . . . . . . . . . . . . . . . . . . CSI5126 . Algorithms in bioinformatics . . . . . . . Space complexity is O ( n k ) (memory cells) For n = 100 and k = 5, n k = 10 10 For n = 100 and k = 10, n k = 10 20 Time complexity is O ( 2 k n k ) Say k = 2 (pairwise) and n = 100 takes 0.2 millisecond For k = 5, n = 100 takes ∼ 26 hours
. Exact . . . . . . . . . Preamble SOP Progressive . Benchmarks Recent methods Preamble SOP Exact Progressive Benchmarks Recent methods Remarks Recall that peta- (P) = 10 15 , tera- (T) 10 12 , giga- (G) 10 9 Given: k sequences of approximately the same length, n Marcel Turcotte . . . . . . . . . . . . . . . . . . . . . . . . . . . . . CSI5126 . Algorithms in bioinformatics Space complexity is O ( n k ) (memory cells) For n = 100 and k = 5, n k = 10 10 For n = 100 and k = 10, n k = 10 20 Time complexity is O ( 2 k n k ) Say k = 2 (pairwise) and n = 100 takes 0.2 millisecond For k = 5, n = 100 takes ∼ 26 hours For k = 10, n = 100 takes ∼ 16 million years!
. Benchmarks . . . . Preamble SOP Exact Progressive Benchmarks Recent methods Preamble SOP Exact Progressive Recent methods . What’s next ? The exact algorithm cannot be applied; prohibitive space and time complexity. What can be done? Use a difgerent optimization technique , something else than dynamic programming; Suggestions? Genetic algorithms SAGA (Notredame and Higgins 1996) Branch-and-bound MSA (Gupta et al 1995), DCA (Stoye et al 1997) Solve a simpler problem: Progressive sequence alignment problem; most widely used approach. Marcel Turcotte . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . CSI5126 . Algorithms in bioinformatics
. Recent methods . . . Preamble SOP Exact Progressive Benchmarks Recent methods Preamble SOP Exact Progressive Benchmarks What’s next ? . The exact algorithm cannot be applied; prohibitive space and time complexity. What can be done? Use a difgerent optimization technique , something else than dynamic programming; Suggestions? Genetic algorithms SAGA (Notredame and Higgins 1996) Branch-and-bound MSA (Gupta et al 1995), DCA (Stoye et al 1997) Solve a simpler problem: Progressive sequence alignment problem; most widely used approach. Marcel Turcotte . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . CSI5126 . Algorithms in bioinformatics
. Benchmarks . . . . Preamble SOP Exact Progressive Benchmarks Recent methods Preamble SOP Exact Progressive Recent methods . What’s next ? The exact algorithm cannot be applied; prohibitive space and time complexity. What can be done? Use a difgerent optimization technique , something else than dynamic programming; Suggestions? Genetic algorithms SAGA (Notredame and Higgins 1996) Branch-and-bound MSA (Gupta et al 1995), DCA (Stoye et al 1997) Solve a simpler problem: Progressive sequence alignment problem; most widely used approach. Marcel Turcotte . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . CSI5126 . Algorithms in bioinformatics
. Benchmarks . . . . Preamble SOP Exact Progressive Benchmarks Recent methods Preamble SOP Exact Progressive Recent methods . What’s next ? The exact algorithm cannot be applied; prohibitive space and time complexity. What can be done? Use a difgerent optimization technique , something else than dynamic programming; Suggestions? Genetic algorithms SAGA (Notredame and Higgins 1996) Branch-and-bound MSA (Gupta et al 1995), DCA (Stoye et al 1997) Solve a simpler problem: Progressive sequence alignment problem; most widely used approach. Marcel Turcotte . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . CSI5126 . Algorithms in bioinformatics
. Benchmarks . . . . Preamble SOP Exact Progressive Benchmarks Recent methods Preamble SOP Exact Progressive Recent methods . What’s next ? The exact algorithm cannot be applied; prohibitive space and time complexity. What can be done? Use a difgerent optimization technique , something else than dynamic programming; Suggestions? Genetic algorithms SAGA (Notredame and Higgins 1996) Branch-and-bound MSA (Gupta et al 1995), DCA (Stoye et al 1997) Solve a simpler problem: Progressive sequence alignment problem; most widely used approach. Marcel Turcotte . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . CSI5126 . Algorithms in bioinformatics
. Benchmarks . . . . Preamble SOP Exact Progressive Benchmarks Recent methods Preamble SOP Exact Progressive Recent methods . What’s next ? The exact algorithm cannot be applied; prohibitive space and time complexity. What can be done? Use a difgerent optimization technique , something else than dynamic programming; Suggestions? Genetic algorithms SAGA (Notredame and Higgins 1996) Branch-and-bound MSA (Gupta et al 1995), DCA (Stoye et al 1997) Solve a simpler problem: Progressive sequence alignment problem; most widely used approach. Marcel Turcotte . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . CSI5126 . Algorithms in bioinformatics
. Recent methods . . . Preamble SOP Exact Progressive Benchmarks Recent methods Preamble SOP Exact Progressive Benchmarks What’s next ? . The exact algorithm cannot be applied; prohibitive space and time complexity. What can be done? Use a difgerent optimization technique , something else than dynamic programming; Suggestions? Genetic algorithms SAGA (Notredame and Higgins 1996) Branch-and-bound MSA (Gupta et al 1995), DCA (Stoye et al 1997) Solve a simpler problem: Progressive sequence alignment problem; most widely used approach. Marcel Turcotte . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . CSI5126 . Algorithms in bioinformatics
. Benchmarks . . . . Preamble SOP Exact Progressive Benchmarks Recent methods Preamble SOP Exact Progressive Recent methods . What’s next ? The exact algorithm cannot be applied; prohibitive space and time complexity. What can be done? Use a difgerent optimization technique , something else than dynamic programming; Suggestions? Genetic algorithms SAGA (Notredame and Higgins 1996) Branch-and-bound MSA (Gupta et al 1995), DCA (Stoye et al 1997) Solve a simpler problem: Progressive sequence alignment problem; most widely used approach. Marcel Turcotte . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . CSI5126 . Algorithms in bioinformatics
. Recent methods . . . . . . . Preamble SOP Exact Progressive Benchmarks Preamble . SOP Exact Progressive Benchmarks Recent methods Progressive alignment methods Idea. 1. Two sequences are chosen and aligned by standard dynamic programming algorithm 2. A third sequence is chosen and aligned to the fjrst alignment 3. Iterate until all sequences have been aligned Marcel Turcotte . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . CSI5126 . Algorithms in bioinformatics ⇒ Most commonly used approach.
. Recent methods . . . . . . . Preamble SOP Exact Progressive Benchmarks Preamble . SOP Exact Progressive Benchmarks Recent methods Progressive Aligments P. Hogeweg and B. Hesper. The alignment of sets of sequences and the construction of phyletic trees: an integrated method. J Mol Evol , 20(2):175–186, 1984. D. G. Higgins and P. M. Sharp. Clustal: a package for performing multiple sequence alignment on a microcomputer. Gene , 73(1):237–44, Dec 1988. Marcel Turcotte . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . CSI5126 . Algorithms in bioinformatics
. Recent methods . . . . . . . Preamble SOP Exact Progressive Benchmarks Preamble . SOP Exact Progressive Benchmarks Recent methods Remarks (digression) Publications are the currency of academia! The number of citations demonstrates the impact of the work in the fjeld. As of 2018-10-03, Des Higgins, the author of Clustal, has 125,800 citations (Scopus, 164,298 citations on Google Scholar)! Marcel Turcotte . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . CSI5126 . Algorithms in bioinformatics
. Benchmarks . . . . . . . . Preamble SOP Exact Progressive Recent methods . Preamble SOP Exact Progressive Benchmarks Recent methods Progressive alignment i and j 2. Build a guide tree 3. From the deepest node up to the root build all the pairwise partial alignments (bottom-up) Marcel Turcotte . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . CSI5126 . Algorithms in bioinformatics 1. Calculate d i , j , distance between sequences i and j , for all
. . . . . .. . . . . . . .. . . . . . . .. . . . . . . .. . . .. . . . . .
. SOP . . . . . . . . . . Preamble Exact . Progressive Benchmarks Recent methods Preamble SOP Exact Progressive Benchmarks Recent methods CLUSTALW Marcel Turcotte . . . . . . . . . . . . . . . . . . . . . . . . . . . . CSI5126 . Algorithms in bioinformatics YDGGPAVEAL YDGGPEAL FEGGPILVEAL FDGGILVQAV YEGGAVVQAL
. SOP . . . . . . . . . . Preamble Exact . Progressive Benchmarks Recent methods Preamble SOP Exact Progressive Benchmarks Recent methods CLUSTALW Marcel Turcotte . . . . . . . . . . . . . . . . . . . . . . . . . . . . CSI5126 . Algorithms in bioinformatics YDGGPAVEAL YDGGPEAL FEGGPILVEAL FDGGILVQAV YEGGAVVQAL
. SOP . . . . . . . . . . Preamble Exact . Progressive Benchmarks Recent methods Preamble SOP Exact Progressive Benchmarks Recent methods CLUSTALW Marcel Turcotte . . . . . . . . . . . . . . . . . . . . . . . . . . . . CSI5126 . Algorithms in bioinformatics YDGGPAVEAL YDGGP−−EAL FEGGPILVEAL FDGGILVQAV YEGGAVVQAL
. SOP . . . . . . . . . . Preamble Exact . Progressive Benchmarks Recent methods Preamble SOP Exact Progressive Benchmarks Recent methods CLUSTALW Marcel Turcotte . . . . . . . . . . . . . . . . . . . . . . . . . . . . CSI5126 . Algorithms in bioinformatics YDGGPAVEAL YDGGP−−EAL FEGGPILVEAL FDGGILVQAV YEGGAVVQAL
. SOP . . . . . . . . . . Preamble Exact . Progressive Benchmarks Recent methods Preamble SOP Exact Progressive Benchmarks Recent methods CLUSTALW Marcel Turcotte . . . . . . . . . . . . . . . . . . . . . . . . . . . . CSI5126 . Algorithms in bioinformatics YDGGPAVEAL YDGGP−−EAL FEGGPILVEAL FDGGILVQAV YEGGAVVQAL
. SOP . . . . . . . . . . Preamble Exact . Progressive Benchmarks Recent methods Preamble SOP Exact Progressive Benchmarks Recent methods CLUSTALW Marcel Turcotte . . . . . . . . . . . . . . . . . . . . . . . . . . . . CSI5126 . Algorithms in bioinformatics YDGGPAVEAL YDGGP−−EAL FEGGPILVEAL FDGGILVQAV YEGGAVVQAL
. SOP . . . . . . . . . . Preamble Exact . Progressive Benchmarks Recent methods Preamble SOP Exact Progressive Benchmarks Recent methods CLUSTALW Marcel Turcotte . . . . . . . . . . . . . . . . . . . . . . . . . . . . CSI5126 . Algorithms in bioinformatics YDGGPA−VEAL YDGGP−−−EAL FEGGPILVEAL FDGGILVQAV YEGGAVVQAL
. SOP . . . . . . . . . . Preamble Exact . Progressive Benchmarks Recent methods Preamble SOP Exact Progressive Benchmarks Recent methods CLUSTALW Marcel Turcotte . . . . . . . . . . . . . . . . . . . . . . . . . . . . CSI5126 . Algorithms in bioinformatics YDGGPA−VEAL YDGGP−−−EAL FEGGPILVEAL FDGGILVQAV YEGGAVVQAL
. SOP . . . . . . . . . . Preamble Exact . Progressive Benchmarks Recent methods Preamble SOP Exact Progressive Benchmarks Recent methods CLUSTALW Marcel Turcotte . . . . . . . . . . . . . . . . . . . . . . . . . . . . CSI5126 . Algorithms in bioinformatics YDGGPA−VEAL YDGGP−−−EAL FEGGPILVEAL FDGGIL−VQAV YEGGAV−VQAL
. SOP . . . . . . . . . . Preamble Exact . Progressive Benchmarks Recent methods Preamble SOP Exact Progressive Benchmarks Recent methods CLUSTALW Marcel Turcotte . . . . . . . . . . . . . . . . . . . . . . . . . . . . CSI5126 . Algorithms in bioinformatics YDGGPA−VEAL YDGGP−−−EAL FEGGPILVEAL FDGGIL−VQAV YEGGAV−VQAL
. Exact . . . . . . . . . Preamble SOP Progressive . Benchmarks Recent methods Preamble SOP Exact Progressive Benchmarks Recent methods Progressive sequence alignment: take 2 Source code available on the course Web site, as well as the Appendix. Marcel Turcotte . . . . . . . . . . . . . . . . . . . . . . . CSI5126 . Algorithms in bioinformatics . . . . . . S1 YDGGPAVEAL S3 S2 YDGGPEAL S5 S4 FEGGPILVEAL S9 S6 FDGGILVQAV S8 S7 YEGGAVVQAL
Sequence vs Sequence , Sequence vs MSA , of the alignment. the sequence. MSA vs MSA ... 0 1 2 3 n − a 1 a a a n 2 3 − 0 b 1 1 b 2 2 ... b m m a 1 , a 2 . . . a n represents a sequence or an alignment When it represents a sequence, the a i are the symbols of When it represents an alignment, the a i are the columns Similarly, for b 1 , b 2 . . . b m . . . . . . .. . . . . . . .. . . . . . . .. . . . . . . .. . . .. . . . . .
. Preamble . . . . . . . . . . . . Exact Progressive Benchmarks Recent methods Preamble SOP Exact Progressive Benchmarks Recent methods Marcel Turcotte . SOP . . . . . . . . . . . . . CSI5126 . Algorithms in bioinformatics . . . . . . . . . . . . . S 1 vs S 2 ... 0 1 2 3 n − a 1 a a a n 2 3 − 0 b 1 1 b 2 2 ... b m m a = S 2 = YDGGPEAL b = S 1 = YDGGPAVEAL
Y 30][ 24][ 18][ 6][ -5][ V [ -42][ -26][ -16][ 20] 26][ 24][ 28] 12][ 1][ D 12] 18][ 24][ 30][ 30][ E [ -48][ -32][ -22][ -11][ 7][ 22] YDGGPAVEAL YDGGP--EAL 30] 18][ 10][ 0][ L [ -60][ -44][ -34][ -23][ -12][ 24][ 0][ 16][ 6][ -6][ A [ -54][ -38][ -28][ -17][ 27] 24][ 22][ 12][ 18][ A [ -36][ -20][ -10][ -4][ Y [ 4][ D [ -12][ -8][ -14][ -20][ -26][ -32] -2][ P [ -30][ -14][ 10][ -6][ -6][ -12][ -18][ -24][ -30][ -36][ -42][ -48] 8][ 0][ [ L A E P G G 14][ 4][ 2][ -8][ 0] -4][ -10][ -16][ -22] 12][ 18][ 24][ 13][ 2][ 6][ G [ -24][ -5][ -11] 1][ 7][ 13][ 19][ 8][ -2][ G [ -18][ . . . . . .. . . . . . . .. . . . . . . .. . . . . . . .. . . .. . . . . .
. . . . . . . . . . . . Preamble Exact . Progressive Benchmarks Recent methods Preamble SOP Exact Progressive Benchmarks Recent methods YDGGP--EAL Marcel Turcotte . SOP . . . . . . . . . . . . . CSI5126 . Algorithms in bioinformatics . . . . . . . . . . . . . S 3 vs S 4 ... 0 1 2 3 n − a 1 a a a n 2 3 − 0 b 1 1 b 2 2 ... b m m a = S 4 = FEGGPILVEAL b = S 3 = YDGGPAVEAL
F 58][ 50][ 26][ 4][ E E [ -62][ -36][ -16][ 14] 25][ 37][ 49][ 59][ 62][ 34][ 60][ 12][ V - [ -54][ -28][ -10][ 12] 24][ 34][ 46][ E 69][ 70][ 46][ 24][ 58][ 59][ A - [ -42][ -16][ L L [ -78][ -52][ -32][ -12][ YDGGPA-VEAL YDGGP---EAL FEGGPILVEAL 85] 63][ 62][ 66][ 68][ 50][ 34][ 10][ 55] 61][ 67][ 61][ 62][ 56][ 50][ 40][ 16][ -6][ A A [ -72][ -46][ -26][ 37] 49][ 2][ 57][ 10] -2][ G G [ -17][ -2][ -14][ -26][ -38][ -50][ -62][ -74] 10][ 22][ 22][ 16][ D D [ -10][ 0][ -12][ -24][ -36][ -48][ -60][ -72][ -84][ -96] 12][ 24][ Y Y [ 27][ 0][ -12][ -24][ -36][ -48][ -60][ -72][ -84][ -96][-108][-120][-132] [ L A E V L I P G G 9][ 34][ 49][ 4][ 34][ 46][ 58][ 70][ 82][ 37][ 36][ 14][ -4][ P P [ -30][ -8][ -20] 58][ 16][ 28][ 40][ 52][ 64][ 42][ 20][ 2][ G G [ -24][ 1][ -11][ -23][ -35][ -47] 13][ 25][ . . . . . .. . . . . . . .. . . . . . . .. . . . . . . .. . . .. . . . . .
. Preamble . . . . . . . . . . . . Exact Progressive Benchmarks Recent methods Preamble SOP Exact Progressive Benchmarks Recent methods Marcel Turcotte . SOP . . . . . . . . . . . . . CSI5126 . Algorithms in bioinformatics . . . . . . . . . . . . . S 5 vs S 6 ... 0 1 2 3 n − a 1 a a a n 2 3 − 0 b 1 1 b 2 2 ... b m m a = S 6 = YEGGAVVQAL b = S 5 = FDGGILVQAV
Y V [ -42][ -29][ -20][ 10] 14][ 19][ 25][ 17][ 8][ 2][ -9][ 6] -4][ 8][ 14][ 20][ 21][ E 8][ -3][ L [ -36][ -23][ -14][ -6] Q [ -48][ -35][ -26][ -15][ 2][ 6][ 25] YEGGAVVQAL FDGGILVQAV 33] 25][ 17][ 9][ 2][ -8][ V [ -60][ -47][ -38][ -27][ -16][ 31][ 11][ 23][ 13][ 5][ -2][ A [ -54][ -41][ -32][ -21][ -10][ 17] 23][ 29][ 19][ 0][ 13][ 12][ -6][ -12][ -18][ -24][ -30][ -36][ -42][ -48][ -54][ -60] 10][ 1][ D [ -12][ -5][ -11][ -17][ -23][ -29][ -35][ -41][ -47] 18][ 7][ -6][ F [ 0][ -2][ [ L A Q V V A G G 4][ 1][ -8][ -14][ -20][ -26][ -32][ -38] 20][ 19][ 14][ 3][ G [ -18][ I [ -30][ -17][ -4][ -10][ -16] 2][ 8][ 14][ -8][ 9][ -2][ G [ -24][ -11][ -9][ -15][ -21][ -27] -3][ 3][ 9][ 15][ 4][ -5][ . . . . . .. . . . . . . .. . . . . . . .. . . . . . . .. . . .. . . . . .
. . . . . . . . . . . Preamble SOP Progressive . Benchmarks Recent methods Preamble SOP Exact Progressive Benchmarks Recent methods YEGGAVVQAL YDGGP---EAL YDGGPA-VEAL Marcel Turcotte . Exact . . . . . . . . . . . . . . CSI5126 . Algorithms in bioinformatics . . . . . . . . . . . . . S 7 vs S 8 ... 0 1 2 3 n − a 1 a a a n 2 3 − 0 b 1 1 b 2 2 ... b m m a = S 8 = FDGGILVQAV b = S 7 = FEGGPILVEAL
Recommend
More recommend